Population History and Gene Divergence in Native Mexicans Inferred from 76 Human Exomes
- PMID: 31848607
- PMCID: PMC7086176
- DOI: 10.1093/molbev/msz282
Population History and Gene Divergence in Native Mexicans Inferred from 76 Human Exomes
Abstract
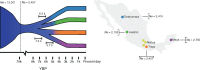
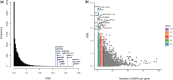
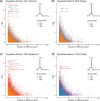
Native American genetic variation remains underrepresented in most catalogs of human genome sequencing data. Previous genotyping efforts have revealed that Mexico's Indigenous population is highly differentiated and substructured, thus potentially harboring higher proportions of private genetic variants of functional and biomedical relevance. Here we have targeted the coding fraction of the genome and characterized its full site frequency spectrum by sequencing 76 exomes from five Indigenous populations across Mexico. Using diffusion approximations, we modeled the demographic history of Indigenous populations from Mexico with northern and southern ethnic groups splitting 7.2 KYA and subsequently diverging locally 6.5 and 5.7 KYA, respectively. Selection scans for positive selection revealed BCL2L13 and KBTBD8 genes as potential candidates for adaptive evolution in Rarámuris and Triquis, respectively. BCL2L13 is highly expressed in skeletal muscle and could be related to physical endurance, a well-known phenotype of the northern Mexico Rarámuri. The KBTBD8 gene has been associated with idiopathic short stature and we found it to be highly differentiated in Triqui, a southern Indigenous group from Oaxaca whose height is extremely low compared to other Native populations.
Keywords: Native Americans; adaptive evolution; demographic inference; exome sequencing.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures