Physiology and Transcriptional Analysis of (p)ppGpp-Related Regulatory Effects in Corynebacterium glutamicum
- PMID: 31849906
- PMCID: PMC6892785
- DOI: 10.3389/fmicb.2019.02769
Physiology and Transcriptional Analysis of (p)ppGpp-Related Regulatory Effects in Corynebacterium glutamicum
Abstract
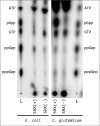
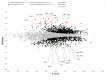
The alarmone species ppGpp and pppGpp are elementary components of bacterial physiology as they both coordinate the bacterial stress response and serve as fine-tuners of general metabolism during conditions of balanced growth. Since the regulation of (p)ppGpp metabolism and the effects of (p)ppGpp on cellular processes are highly complex and show massive differences between bacterial species, the underlying molecular mechanisms have so far only been insufficiently investigated for numerous microorganisms. In this study, (p)ppGpp physiology in the actinobacterial model organism Corynebacterium glutamicum was analyzed by phenotypic characterization and RNAseq-based transcriptome analysis. Total nutrient starvation was identified as the most effective method to induce alarmone production, whereas traditional induction methods such as the addition of serine hydroxamate (SHX) or mupirocin did not show a strong accumulation of (p)ppGpp. The predominant alarmone in C. glutamicum represents guanosine tetraphosphate, whose stress-associated production depends on the presence of the bifunctional RSH enzyme Rel. Interestingly, in addition to ppGpp, another substance yet not identified accumulated strongly under inducing conditions. A C. glutamicum triple mutant (Δrel,ΔrelS,ΔrelH) unable to produce alarmones [(p)ppGpp0 strain] exhibited unstable growth characteristics and interesting features such as an influence of illumination on its physiology, production of amino acids as well as differences in vitamin and carotenoid production. Differential transcriptome analysis using RNAseq provided numerous indications for the molecular basis of the observed phenotype. An evaluation of the (p)ppGpp-dependent transcriptional regulation under total nutrient starvation revealed a complex interplay with the involvement of ribosome-mediated transcriptional attenuation, the stress-responsive sigma factors σB and σH and transcription factors such as McbR, the master regulator of sulfur metabolism. In addition to the differential regulation of genes connected with various cell functions, the transcriptome analysis revealed conserved motifs within the promoter regions of (p)ppGpp-dependently and independently regulated genes. In particular, the representatives of translation-associated genes are both (p)ppGpp-dependent transcriptionally downregulated and show a highly conserved and so far unknown TTTTG motif in the -35 region, which is also present in other actinobacterial genera.
Keywords: (p)ppGpp; Rel; discriminator; promoter; stress response; stringent response.
Copyright © 2019 Ruwe, Persicke, Busche, Müller and Kalinowski.
Figures







References
-
- Baumgart M., Unthan S., Rückert C., Sivalingam J., Grünberger A., Kalinowski J., et al. (2013). Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 79 6006–6015. 10.1128/AEM.01634-13 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

