K5 Capsule and Lipopolysaccharide Are Important in Resistance to T4 Phage Attack in Probiotic E. coli Strain Nissle 1917
- PMID: 31849915
- PMCID: PMC6895014
- DOI: 10.3389/fmicb.2019.02783
K5 Capsule and Lipopolysaccharide Are Important in Resistance to T4 Phage Attack in Probiotic E. coli Strain Nissle 1917
Abstract
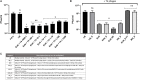
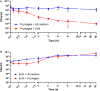
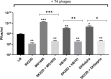
Rapidly growing antibiotic resistance among gastrointestinal pathogens, and the ability of antibiotics to induce the virulence of these pathogens makes it increasingly difficult to rely on antibiotics to treat gastrointestinal infections. The probiotic Escherichia coli strain Nissle 1917 (EcN) is the active component of the pharmaceutical preparation Mutaflor® and has been successfully used in the treatment of gastrointestinal disorders. Gut bacteriophages are dominant players in maintaining the microbial homeostasis in the gut, however, their interaction with incoming probiotic bacteria remains to be at conception. The presence of bacteriophages in the gut makes it inevitable for any probiotic bacteria to be phage resistant, in order to survive and successfully colonize the gut. This study addresses the phage resistance of EcN, specifically against lytic T4 phage infection. From various experiments we could show that (i) EcN is resistant toward T4 phage infection, (ii) EcN's K5 polysaccharide capsule plays a crucial role in T4 phage resistance and (iii) EcN's lipopolysaccharide (LPS) inactivates T4 phages and notably, treatment with the antibiotic polymyxin B which neutralizes the LPS destroyed the phage inactivation ability of isolated LPS from EcN. Combination of these identified properties in EcN was not found in other tested commensal E. coli strains. Our results further indicated that N-acetylglucosamine at the distal end of O6 antigen in EcN's LPS could be the interacting partner with T4 phages. From our findings, we have reported for the first time, the role of EcN's K5 capsule and LPS in its defense against T4 phages. In addition, by inactivating the T4 phages, EcN also protects E. coli K-12 strains from phage infection in tri-culture experiments. Our research highlights phage resistance as an additional safety feature of EcN, a clinically successful probiotic E. coli strain.
Keywords: E. coli Nissle 1917; K5 capsule; Mutaflor; T4 phages; gastrointestinal infections; lipopolysaccharide; phage resistance; probiotics.
Copyright © 2019 Soundararajan, von Bünau and Oelschlaeger.
Figures







References
-
- Arribas B., Rodriguez-Cabezas M. E., Camuesco D., Comalada M., Bailon E., Utrilla P., et al. (2009). A probiotic strain of Escherichia coli, Nissle 1917, given orally exerts local and systemic anti-inflammatory effects in lipopolysaccharide-induced sepsis in mice. Br. J. Pharmacol. 157 1024–1033. 10.1111/j.1476-5381.2009.00270.x - DOI - PMC - PubMed
-
- Beacham I. R., Picken R. N. (1981). On the receptor for bacteriophage T4 in Escherichia coli K12. Curr. Microbiol. 6 291–293. 10.1007/bf01566879 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

