Evolutionary Models for the Diversification of Placental Mammals Across the KPg Boundary
- PMID: 31850081
- PMCID: PMC6896846
- DOI: 10.3389/fgene.2019.01241
Evolutionary Models for the Diversification of Placental Mammals Across the KPg Boundary
Abstract
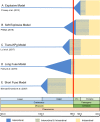
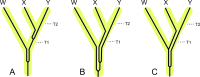
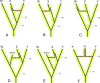
Deciphering the timing of the placental mammal radiation is a longstanding problem in evolutionary biology, but consensus on the tempo and mode of placental diversification remains elusive. Nevertheless, an accurate timetree is essential for understanding the role of important events in Earth history (e.g., Cretaceous Terrestrial Revolution, KPg mass extinction) in promoting the taxonomic and ecomorphological diversification of Placentalia. Archibald and Deutschman described three competing models for the diversification of placental mammals, which are the Explosive, Long Fuse, and Short Fuse Models. More recently, the Soft Explosive Model and Trans-KPg Model have emerged as additional hypotheses for the placental radiation. Here, we review molecular and paleontological evidence for each of these five models including the identification of general problems that can negatively impact divergence time estimates. The Long Fuse Model has received more support from relaxed clock studies than any of the other models, but this model is not supported by morphological cladistic studies that position Cretaceous eutherians outside of crown Placentalia. At the same time, morphological cladistics has a poor track record of reconstructing higher-level relationships among the orders of placental mammals including the results of new pseudoextinction analyses that we performed on the largest available morphological data set for mammals (4,541 characters). We also examine the strengths and weaknesses of different timetree methods (node dating, tip dating, and fossilized birth-death dating) that may now be applied to estimate the timing of the placental radiation. While new methods such as tip dating are promising, they also have problems that must be addressed if these methods are to effectively discriminate among competing hypotheses for placental diversification. Finally, we discuss the complexities of timetree estimation when the signal of speciation times is impacted by incomplete lineage sorting (ILS) and hybridization. Not accounting for ILS results in dates that are older than speciation events. Hybridization, in turn, can result in dates than are younger or older than speciation dates. Disregarding this potential variation in "gene" history across the genome can distort phylogenetic branch lengths and divergence estimates when multiple unlinked genomic loci are combined together in a timetree analysis.
Keywords: KPg boundary; placental radiation; relaxed clocks; timetrees; tip dating.
Copyright © 2019 Springer, Foley, Brady, Gatesy and Murphy.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources

