Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules
- PMID: 31851501
- PMCID: PMC7145764
- DOI: 10.1021/acs.chemrev.9b00440
Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules
Abstract
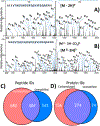
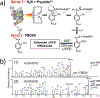
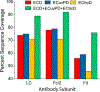
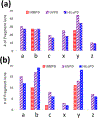
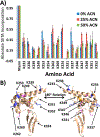
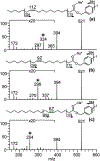
The development of new ion-activation/dissociation methods continues to be one of the most active areas of mass spectrometry owing to the broad applications of tandem mass spectrometry in the identification and structural characterization of molecules. This Review will showcase the impact of ultraviolet photodissociation (UVPD) as a frontier strategy for generating informative fragmentation patterns of ions, especially for biological molecules whose complicated structures, subtle modifications, and large sizes often impede molecular characterization. UVPD energizes ions via absorption of high-energy photons, which allows access to new dissociation pathways relative to more conventional ion-activation methods. Applications of UVPD for the analysis of peptides, proteins, lipids, and other classes of biologically relevant molecules are emphasized in this Review.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

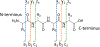
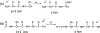






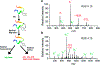

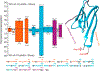
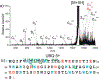

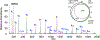








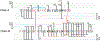


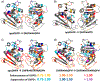




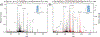

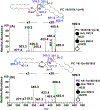
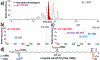

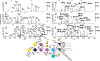
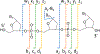


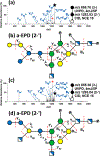


References
-
- Banoub JH; Newton RP; Esmans E; Ewing DF; Mackenzie G Recent development in mass spectrometry for the characterization of nucleosides, nucleotides, oligonucleotides, and nucleic acids. Chem. Rev 2005, 105, 1869–1915. - PubMed
-
- Fabris D A role for the MS analysis of nucleic acids in the post-genomics age. J. Am. Soc. Mass Spectrom 2010, 21, 1–13. - PubMed
-
- Schürch S Characterization of nucleic acids by tandem mass spectrometry - The second decade (2004–2013): From DNA to RNA and modified sequences. Mass Spectrom. Rev 2016, 35, 483–523. - PubMed
-
- Beck JL; Colgrave MO; Ralph SF; Sheil MM Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom. Rev 2001, 20, 61–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

