Cellular IP6 Levels Limit HIV Production while Viruses that Cannot Efficiently Package IP6 Are Attenuated for Infection and Replication
- PMID: 31851928
- PMCID: PMC6931105
- DOI: 10.1016/j.celrep.2019.11.050
Cellular IP6 Levels Limit HIV Production while Viruses that Cannot Efficiently Package IP6 Are Attenuated for Infection and Replication
Abstract
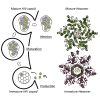
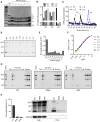
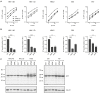
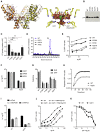
HIV-1 hijacks host proteins to promote infection. Here we show that HIV is also dependent upon the host metabolite inositol hexakisphosphate (IP6) for viral production and primary cell replication. HIV-1 recruits IP6 into virions using two lysine rings in its immature hexamers. Mutation of either ring inhibits IP6 packaging and reduces viral production. Loss of IP6 also results in virions with highly unstable capsids, leading to a profound loss of reverse transcription and cell infection. Replacement of one ring with a hydrophobic isoleucine core restores viral production, but IP6 incorporation and infection remain impaired, consistent with an independent role for IP6 in stable capsid assembly. Genetic knockout of biosynthetic kinases IPMK and IPPK reveals that cellular IP6 availability limits the production of diverse lentiviruses, but in the absence of IP6, HIV-1 packages IP5 without loss of infectivity. Together, these data suggest that IP6 is a critical cofactor for HIV-1 replication.
Keywords: AIDS; HIV; IP6; IPMK; IPPK; capsid; inositol hexakisphosphate; virus.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Azevedo C., Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 2006;1:2416–2422. - PubMed
-
- Brissault B., Kichler A., Guis C., Leborgne C., Danos O., Cheradame H. Synthesis of linear polyethylenimine derivatives for DNA transfection. Bioconjug. Chem. 2003;14:581–587. - PubMed
-
- Bush D.L., Vogt V.M. In vitro assembly of retroviruses. Annu. Rev. Virol. 2014;1:561–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

