Laboratory Analysis of an Outbreak of Candida auris in New York from 2016 to 2018: Impact and Lessons Learned
- PMID: 31852764
- PMCID: PMC7098748
- DOI: 10.1128/JCM.01503-19
Laboratory Analysis of an Outbreak of Candida auris in New York from 2016 to 2018: Impact and Lessons Learned
Abstract
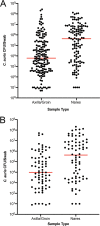
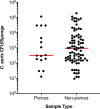
Candida auris is a multidrug-resistant yeast which has emerged in health care facilities worldwide; however, little is known about identification methods, patient colonization, environmental survival, spread, and drug resistance. Colonization on both biotic (patients) and abiotic (health care objects) surfaces, along with travel, appear to be the major factors for the spread of this pathogen across the globe. In this investigation, we present laboratory findings from an ongoing C. auris outbreak in New York (NY) from August 2016 through 2018. A total of 540 clinical isolates, 11,035 patient surveillance specimens, and 3,672 environmental surveillance samples were analyzed. Laboratory methods included matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for yeast isolate identification, real-time PCR for rapid surveillance sample screening, culture on selective/nonselective media for recovery of C. auris and other yeasts from surveillance samples, antifungal susceptibility testing to determine the C. auris resistance profile, and Sanger sequencing of the internal transcribed spacer (ITS) and D1/D2 regions of the ribosomal gene for C. auris genotyping. Results included (a) identification and confirmation of C. auris in 413 clinical isolates and 931 patient surveillance isolates as well as identification of 277 clinical cases and 350 colonized cases from 151 health care facilities, including 59 hospitals, 92 nursing homes, 1 long-term acute care hospital (LTACH), and 2 hospices, (b) successful utilization of an in-house developed C. auris real-time PCR assay for the rapid screening of patient and environmental surveillance samples, (c) demonstration of relatively heavier colonization of C. auris in nares than in the axilla/groin, and (d) predominance of the South Asia clade I with intrinsic resistance to fluconazole and elevated MIC to voriconazole (81%), amphotericin B (61%), flucytosine (5FC) (3%), and echinocandins (1%). These findings reflect greater regional prevalence and incidence of C. auris and the deployment of better detection tools in an unprecedented outbreak.
Keywords: Candida auris; antifungals; molecular biology; mycology; phylogenetics.
Copyright © 2020 American Society for Microbiology.
Figures

Comment in
-
Analysis of a Candida auris Outbreak Provides New Insights into an Emerging Pathogen.J Clin Microbiol. 2020 Mar 25;58(4):e02083-19. doi: 10.1128/JCM.02083-19. Print 2020 Mar 25. J Clin Microbiol. 2020. PMID: 31996439 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

