Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity
- PMID: 31852826
- PMCID: PMC6955338
- DOI: 10.1073/pnas.1913690116
Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity
Abstract
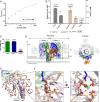
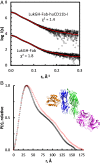
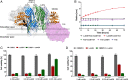
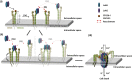
Host-pathogen interactions are central to understanding microbial pathogenesis. The staphylococcal pore-forming cytotoxins hijack important immune molecules but little is known about the underlying molecular mechanisms of cytotoxin-receptor interaction and host specificity. Here we report the structures of a staphylococcal pore-forming cytotoxin, leukocidin GH (LukGH), in complex with its receptor (the α-I domain of complement receptor 3, CD11b-I), both for the human and murine homologs. We observe 2 binding interfaces, on the LukG and the LukH protomers, and show that human CD11b-I induces LukGH oligomerization in solution. LukGH binds murine CD11b-I weakly and is inactive toward murine neutrophils. Using a LukGH variant engineered to bind mouse CD11b-I, we demonstrate that cytolytic activity does not only require binding but also receptor-dependent oligomerization. Our studies provide an unprecedented insight into bicomponent leukocidin-host receptor interaction, enabling the development of antitoxin approaches and improved animal models to explore these approaches.
Keywords: host–pathogen interaction; integrin; leukocidin; pore forming toxins; receptor recognition.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: A.B. and H.R. are employees of X4 Pharmaceuticals GmbH, the legal successor of Arsanis Biosciences GmbH, which has developed an antileukocidin-GH antibody.
Figures


References
-
- Yamashita D., et al. , Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins. Nat. Commun. 5, 4897 (2014). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

