Genomic and Phenotypic Characterization of a Broad Panel of Patient-Derived Xenografts Reflects the Diversity of Glioblastoma
- PMID: 31852831
- PMCID: PMC7056576
- DOI: 10.1158/1078-0432.CCR-19-0909
Genomic and Phenotypic Characterization of a Broad Panel of Patient-Derived Xenografts Reflects the Diversity of Glioblastoma
Abstract
Purpose: Glioblastoma is the most frequent and lethal primary brain tumor. Development of novel therapies relies on the availability of relevant preclinical models. We have established a panel of 96 glioblastoma patient-derived xenografts (PDX) and undertaken its genomic and phenotypic characterization.
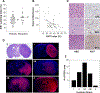
Experimental design: PDXs were established from glioblastoma, IDH-wildtype (n = 93), glioblastoma, IDH-mutant (n = 2), diffuse midline glioma, H3 K27M-mutant (n = 1), and both primary (n = 60) and recurrent (n = 34) tumors. Tumor growth rates, histopathology, and treatment response were characterized. Integrated molecular profiling was performed by whole-exome sequencing (WES, n = 83), RNA-sequencing (n = 68), and genome-wide methylation profiling (n = 76). WES data from 24 patient tumors was compared with derivative models.
Results: PDXs recapitulate many key phenotypic and molecular features of patient tumors. Orthotopic PDXs show characteristic tumor morphology and invasion patterns, but largely lack microvascular proliferation and necrosis. PDXs capture common and rare molecular drivers, including alterations of TERT, EGFR, PTEN, TP53, BRAF, and IDH1, most at frequencies comparable with human glioblastoma. However, PDGFRA amplification was absent. RNA-sequencing and genome-wide methylation profiling demonstrated broad representation of glioblastoma molecular subtypes. MGMT promoter methylation correlated with increased survival in response to temozolomide. WES of 24 matched patient tumors showed preservation of most genetic driver alterations, including EGFR amplification. However, in four patient-PDX pairs, driver alterations were gained or lost on engraftment, consistent with clonal selection.
Conclusions: Our PDX panel captures the molecular heterogeneity of glioblastoma and recapitulates many salient genetic and phenotypic features. All models and genomic data are openly available to investigators.
©2019 American Association for Cancer Research.
Figures
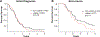

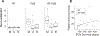

References
-
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006;9(5):391–403 doi 10.1016/j.ccr.2006.03.030. - DOI - PubMed
-
- Schulte A, Gunther HS, Martens T, Zapf S, Riethdorf S, Wulfing C, et al. Glioblastoma stem-like cell lines with either maintenance or loss of high-level EGFR amplification, generated via modulation of ligand concentration. Clin Cancer Res 2012;18(7):1901–13 doi 10.1158/1078-0432.CCR-11-3084. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

