Automated acquisition of explainable knowledge from unannotated histopathology images
- PMID: 31852890
- PMCID: PMC6920352
- DOI: 10.1038/s41467-019-13647-8
Automated acquisition of explainable knowledge from unannotated histopathology images
Abstract
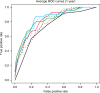
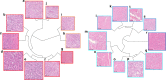
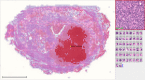
Deep learning algorithms have been successfully used in medical image classification. In the next stage, the technology of acquiring explainable knowledge from medical images is highly desired. Here we show that deep learning algorithm enables automated acquisition of explainable features from diagnostic annotation-free histopathology images. We compare the prediction accuracy of prostate cancer recurrence using our algorithm-generated features with that of diagnosis by expert pathologists using established criteria on 13,188 whole-mount pathology images consisting of over 86 billion image patches. Our method not only reveals findings established by humans but also features that have not been recognized, showing higher accuracy than human in prognostic prediction. Combining both our algorithm-generated features and human-established criteria predicts the recurrence more accurately than using either method alone. We confirm robustness of our method using external validation datasets including 2276 pathology images. This study opens up fields of machine learning analysis for discovering uncharted knowledge.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
The emerging role of artificial intelligence in the reporting of prostate pathology.Pathology. 2021 Aug;53(5):565-567. doi: 10.1016/j.pathol.2021.04.002. Epub 2021 Jun 6. Pathology. 2021. PMID: 34108086 No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

