Characterization of protein, long noncoding RNA and microRNA signatures in extracellular vesicles derived from resting and degranulated mast cells
- PMID: 31853339
- PMCID: PMC6913652
- DOI: 10.1080/20013078.2019.1697583
Characterization of protein, long noncoding RNA and microRNA signatures in extracellular vesicles derived from resting and degranulated mast cells
Abstract
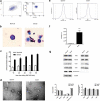
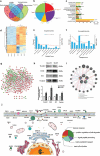
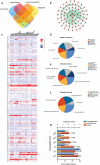
Mast cells (MCs) are known to participate in a variety of patho-physiological processes depending largely on the intragranular mediators and the production of cytokines and chemokines during degranulation. Recently, extracellular vesicles (EVs) have been implicated important functions for MCs, but the components of MC-derived EVs have not yet been well-characterized. In this study, we aimed to identify signatures of proteins, long non-coding RNAs (lncRNAs), and microRNAs (miRNAs) in EVs derived from resting (Rest-EV) and degranulated (Sti-EV) MCs by differential ultracentrifugation. Using tandem mass tag (TMT)-based quantitative proteomics technology and RNA sequencing, we identified a total of 1988 proteins, 397 lncRNAs, and 272 miRNAs in Rest-EV and Sti-EV. The proteins include common EVs markers (cytoskeletal proteins), MCs markers (FcεRI and tryptase), and some preformed MCs mediators (lysosomal enzymes) as well. The global expression profiles of lncRNAs and miRNAs identified, for the first time, from Rest-EV and Sti-EV, strongly suggest a potential regulatory function of MC-derived EVs. We have also performed Western blotting and qRT-PCR analysis to further verify some of the proteins, lncRNAs, and miRNAs identified from Rest-EV and Sti-EV. Our findings will help to elucidate the functions of MC-derived EVs, and provide a reference dataset for future translational studies involving MC-derived EVs.
Keywords: EVs; Mast cells; lncRNAs; miRNAs; proteomics.
© 2019 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group on behalf of The International Society for Extracellular Vesicles.
Conflict of interest statement
No potential conflicts of interest were reported by the authors.
Figures



References
-
- Li Z, Liu S, Xu J, et al. Adult connective tissue-resident mast cells originate from late erythro-myeloid progenitors. Immunity. 2018. October 16;49(4):640–653 e5. PubMed PMID: 30332630. - PubMed
-
- Merluzzi S, Betto E, Ceccaroni AA, et al. Mast cells, basophils and B cell connection network. Mol Immunol. 2015. January;63(1):94–103. PubMed PMID: 24671125. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

