De novo design of a homo-trimeric amantadine-binding protein
- PMID: 31854299
- PMCID: PMC6922598
- DOI: 10.7554/eLife.47839
De novo design of a homo-trimeric amantadine-binding protein
Abstract
The computational design of a symmetric protein homo-oligomer that binds a symmetry-matched small molecule larger than a metal ion has not yet been achieved. We used de novo protein design to create a homo-trimeric protein that binds the C3 symmetric small molecule drug amantadine with each protein monomer making identical interactions with each face of the small molecule. Solution NMR data show that the protein has regular three-fold symmetry and undergoes localized structural changes upon ligand binding. A high-resolution X-ray structure reveals a close overall match to the design model with the exception of water molecules in the amantadine binding site not included in the Rosetta design calculations, and a neutron structure provides experimental validation of the computationally designed hydrogen-bond networks. Exploration of approaches to generate a small molecule inducible homo-trimerization system based on the design highlight challenges that must be overcome to computationally design such systems.
Keywords: E. coli; Rosetta; amantadine; de novo protein design; molecular biophysics; structural biology; symmetry.
© 2019, Park et al.
Conflict of interest statement
JP, SB, KW, GO, DB JP, SEB, KYW, GO, and DB have filed a provisional application based for "Amantadine Binding Protein" (Application # 62/834,592). BS, AM, WD, NS, MC, DM No competing interests declared
Figures


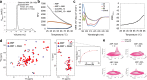


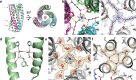

References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Arzt S, Campbell JW, Harding MM, Hao Q, Helliwell JR. LSCALE-the new normalization, scaling and absorption correction program in the Daresbury laue software suite. Journal of Applied Crystallography. 1999;32:554–562. doi: 10.1107/S0021889898015350. - DOI
-
- Boyken SE, Chen Z, Groves B, Langan RA, Oberdorfer G, Ford A, Gilmore JM, Xu C, DiMaio F, Pereira JH, Sankaran B, Seelig G, Zwart PH, Baker D. De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science. 2016;352:680–687. doi: 10.1126/science.aad8865. - DOI - PMC - PubMed

