Variability of mitochondrial ORFans hints at possible differences in the system of doubly uniparental inheritance of mitochondria among families of freshwater mussels (Bivalvia: Unionida)
- PMID: 31856711
- PMCID: PMC6923999
- DOI: 10.1186/s12862-019-1554-5
Variability of mitochondrial ORFans hints at possible differences in the system of doubly uniparental inheritance of mitochondria among families of freshwater mussels (Bivalvia: Unionida)
Abstract
Background: Supernumerary ORFan genes (i.e., open reading frames without obvious homology to other genes) are present in the mitochondrial genomes of gonochoric freshwater mussels (Bivalvia: Unionida) showing doubly uniparental inheritance (DUI) of mitochondria. DUI is a system in which distinct female-transmitted and male-transmitted mitotypes coexist in a single species. In families Unionidae and Margaritiferidae, the transition from dioecy to hermaphroditism and the loss of DUI appear to be linked, and this event seems to affect the integrity of the ORFan genes. These observations led to the hypothesis that the ORFans have a role in DUI and/or sex determination. Complete mitochondrial genome sequences are however scarce for most families of freshwater mussels, therefore hindering a clear localization of DUI in the various lineages and a comprehensive understanding of the influence of the ORFans on DUI and sexual systems. Therefore, we sequenced and characterized eleven new mitogenomes from poorly sampled freshwater mussel families to gather information on the evolution and variability of the ORFan genes and their protein products.
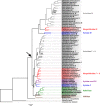
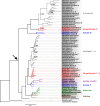
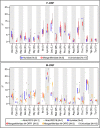
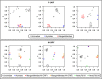
Results: We obtained ten complete plus one almost complete mitogenome sequence from ten representative species (gonochoric and hermaphroditic) of families Margaritiferidae, Hyriidae, Mulleriidae, and Iridinidae. ORFan genes are present only in DUI species from Margaritiferidae and Hyriidae, while non-DUI species from Hyriidae, Iridinidae, and Mulleriidae lack them completely, independently of their sexual system. Comparisons among the proteins translated from the newly characterized ORFans and already known ones provide evidence of conserved structures, as well as family-specific features.
Conclusions: The ORFan proteins show a comparable organization of secondary structures among different families of freshwater mussels, which supports a conserved physiological role, but also have distinctive family-specific features. Given this latter observation and the fact that the ORFans can be either highly mutated or completely absent in species that secondarily lost DUI depending on their respective family, we hypothesize that some aspects of the connection among ORFans, sexual systems, and DUI may differ in the various lineages of unionids.
Keywords: Doubly uniparental inheritance of mitochondrial DNA; Evolution of protein structures and functions; Freshwater mussels; Mitochondria and sexual systems; Mitochondrial ORFan genes; mtDNA sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
In silico analyses of mitochondrial ORFans in freshwater mussels (Bivalvia: Unionoida) provide a framework for future studies of their origin and function.BMC Genomics. 2016 Aug 9;17:597. doi: 10.1186/s12864-016-2986-6. BMC Genomics. 2016. PMID: 27507266 Free PMC article.
-
Evolution of sex-dependent mtDNA transmission in freshwater mussels (Bivalvia: Unionida).Sci Rep. 2017 May 8;7(1):1551. doi: 10.1038/s41598-017-01708-1. Sci Rep. 2017. PMID: 28484275 Free PMC article.
-
The complete mitochondrial genome of the hermaphroditic freshwater mussel Anodonta cygnea (Bivalvia: Unionidae): in silico analyses of sex-specific ORFs across order Unionoida.BMC Genomics. 2018 Mar 27;19(1):221. doi: 10.1186/s12864-018-4583-3. BMC Genomics. 2018. PMID: 29587633 Free PMC article.
-
The exceptional mitochondrial DNA system of the mussel family Mytilidae.Genes Genet Syst. 2000 Dec;75(6):313-8. doi: 10.1266/ggs.75.313. Genes Genet Syst. 2000. PMID: 11280005 Review.
-
The unusual system of doubly uniparental inheritance of mtDNA: isn't one enough?Trends Genet. 2007 Sep;23(9):465-74. doi: 10.1016/j.tig.2007.05.011. Epub 2007 Aug 6. Trends Genet. 2007. PMID: 17681397 Review.
Cited by
-
A tale of two paths: The evolution of mitochondrial recombination in bivalves with doubly uniparental inheritance.J Hered. 2023 May 25;114(3):199-206. doi: 10.1093/jhered/esad004. J Hered. 2023. PMID: 36897956 Free PMC article.
-
ORFans in Mitochondrial Genomes of Marine Polychaete Polydora.Genome Biol Evol. 2023 Dec 1;15(12):evad219. doi: 10.1093/gbe/evad219. Genome Biol Evol. 2023. PMID: 38019573 Free PMC article.
-
Mitonuclear Sex Determination? Empirical Evidence from Bivalves.Mol Biol Evol. 2023 Nov 3;40(11):msad240. doi: 10.1093/molbev/msad240. Mol Biol Evol. 2023. PMID: 37935058 Free PMC article.
-
Did doubly uniparental inheritance (DUI) of mtDNA originate as a cytoplasmic male sterility (CMS) system?Bioessays. 2022 Apr;44(4):e2100283. doi: 10.1002/bies.202100283. Epub 2022 Feb 16. Bioessays. 2022. PMID: 35170770 Free PMC article.
-
The complete mitogenome of the inarticulate brachiopod Glottidia pyramidata reveals insights into gene order variation, deviant ATP8 and mtORFans in the Brachiopoda.Mitochondrial DNA B Resour. 2021 Aug 18;6(9):2701-2703. doi: 10.1080/23802359.2021.1966342. eCollection 2021. Mitochondrial DNA B Resour. 2021. PMID: 34435125 Free PMC article.
References
-
- Zouros E. Biparental inheritance through Uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol. 2013;40:1–31. doi: 10.1007/s11692-012-9195-2. - DOI
-
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 2010;10:50. doi: 10.1186/1471-2148-10-50. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

