Edematous severe acute malnutrition is characterized by hypomethylation of DNA
- PMID: 31857576
- PMCID: PMC6923441
- DOI: 10.1038/s41467-019-13433-6
Edematous severe acute malnutrition is characterized by hypomethylation of DNA
Abstract
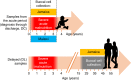
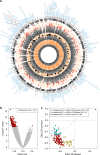
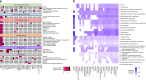
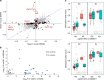
Edematous severe acute childhood malnutrition (edematous SAM or ESAM), which includes kwashiorkor, presents with more overt multi-organ dysfunction than non-edematous SAM (NESAM). Reduced concentrations and methyl-flux of methionine in 1-carbon metabolism have been reported in acute, but not recovered, ESAM, suggesting downstream DNA methylation changes could be relevant to differences in SAM pathogenesis. Here, we assess genome-wide DNA methylation in buccal cells of 309 SAM children using the 450 K microarray. Relative to NESAM, ESAM is characterized by multiple significantly hypomethylated loci, which is not observed among SAM-recovered adults. Gene expression and methylation show both positive and negative correlation, suggesting a complex transcriptional response to SAM. Hypomethylated loci link to disorders of nutrition and metabolism, including fatty liver and diabetes, and appear to be influenced by genetic variation. Our epigenetic findings provide a potential molecular link to reported aberrant 1-carbon metabolism in ESAM and support consideration of methyl-group supplementation in ESAM.
Conflict of interest statement
J.W.B. and X.W. are fulltime employees of Illumina Inc., but all work relevant to the current study was performed whilst they were employees of Baylor College of Medicine. The remaining authors have no competing interests to declare.
Figures

References
-
- M. H. N. Golden, M. H. N. & Golden, B. E. Severe Malnutrition Vol. 3. Oxford Textbook of Medicine. 1278−1296 (Oxford, 1996).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

