Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance
- PMID: 31860775
- PMCID: PMC7036369
- DOI: 10.1111/mpp.12897
Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance
Abstract
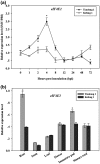
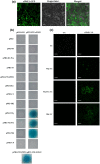
Soybean mosaic virus (SMV), a potyvirus, is the most prevalent and destructive viral pathogen in soybean-planting regions of China. Moreover, other potyviruses, including bean common mosaic virus (BCMV) and watermelon mosaic virus (WMV), also threaten soybean farming. The eukaryotic translation initiation factor 4E (eIF4E) plays a critical role in controlling resistance/susceptibility to potyviruses in plants. In the present study, much higher SMV-induced eIF4E1 expression levels were detected in a susceptible soybean cultivar when compared with a resistant cultivar, suggesting the involvement of eIF4E1 in the response to SMV by the susceptible cultivar. Yeast two-hybrid and bimolecular fluorescence complementation assays showed that soybean eIF4E1 interacted with SMV VPg in the nucleus and with SMV NIa-Pro/NIb in the cytoplasm, revealing the involvement of VPg, NIa-Pro, and NIb in SMV infection and multiplication. Furthermore, transgenic soybeans silenced for eIF4E were produced using an RNA interference approach. Through monitoring for viral symptoms and viral titers, robust and broad-spectrum resistance was confirmed against five SMV strains (SC3/7/15/18 and SMV-R), BCMV, and WMV in the transgenic plants. Our findings represent fresh insights for investigating the mechanism underlying eIF4E-mediated resistance in soybean and also suggest an effective alternative for breeding soybean with broad-spectrum viral resistance.
Keywords: Potyvirus; Soybean mosaic virus; Agrobacterium-mediated transformation; RNA interference; broad-spectrum resistance; eIF4E; soybean.
© 2019 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest.
Figures



Similar articles
-
RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean.Plant Cell Rep. 2018 Jan;37(1):103-114. doi: 10.1007/s00299-017-2186-0. Epub 2017 Jul 29. Plant Cell Rep. 2018. PMID: 28756582
-
Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato.PLoS One. 2011;6(12):e29595. doi: 10.1371/journal.pone.0029595. Epub 2011 Dec 29. PLoS One. 2011. PMID: 22242134 Free PMC article.
-
Robust RNAi-mediated resistance to infection of seven potyvirids in soybean expressing an intron hairpin NIb RNA.Transgenic Res. 2017 Oct;26(5):665-676. doi: 10.1007/s11248-017-0041-2. Epub 2017 Aug 24. Transgenic Res. 2017. PMID: 28840434
-
Decades of Genetic Research on Soybean mosaic virus Resistance in Soybean.Viruses. 2022 May 24;14(6):1122. doi: 10.3390/v14061122. Viruses. 2022. PMID: 35746594 Free PMC article. Review.
-
The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection.Viruses. 2020 Jan 8;12(1):77. doi: 10.3390/v12010077. Viruses. 2020. PMID: 31936267 Free PMC article. Review.
Cited by
-
Pathogenicity and genome-wide sequence analysis reveals relationships between soybean mosaic virus strains.Arch Virol. 2022 Feb;167(2):517-529. doi: 10.1007/s00705-021-05271-z. Epub 2022 Jan 13. Arch Virol. 2022. PMID: 35024966 Free PMC article.
-
Development of Comprehensive Serological Techniques for Sensitive, Quantitative and Rapid Detection of Soybean mosaic virus.Int J Mol Sci. 2022 Aug 21;23(16):9457. doi: 10.3390/ijms23169457. Int J Mol Sci. 2022. PMID: 36012722 Free PMC article.
-
Interaction Between Translation Initiation Factor eIF4E in Banana and the VPg Protein of Banana Bract Mosaic Virus Reveals Potential Targets for Genome Editing.Mol Biotechnol. 2024 Dec 23. doi: 10.1007/s12033-024-01337-w. Online ahead of print. Mol Biotechnol. 2024. PMID: 39715933
-
Selective Interaction of Sugarcane eIF4E with VPgs from Sugarcane Mosaic Pathogens.Viruses. 2021 Mar 22;13(3):518. doi: 10.3390/v13030518. Viruses. 2021. PMID: 33809985 Free PMC article.
-
RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture.Plants (Basel). 2021 Sep 14;10(9):1914. doi: 10.3390/plants10091914. Plants (Basel). 2021. PMID: 34579446 Free PMC article. Review.
References
-
- Albar, L. , Bangratz‐Reyser, M. , Hébrard, E. , Ndjiondjop, M.N. , Jones, M. and Ghesquière, A. (2006) Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus . Plant Journal, 47, 417–426. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

