Functional Analysis of DNMT3A DNA Methyltransferase Mutations Reported in Patients with Acute Myeloid Leukemia
- PMID: 31861499
- PMCID: PMC7022712
- DOI: 10.3390/biom10010008
Functional Analysis of DNMT3A DNA Methyltransferase Mutations Reported in Patients with Acute Myeloid Leukemia
Abstract
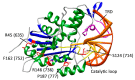
In mammals, DNA methylation is necessary for the maintenance of genomic stability, gene expression regulation, and other processes. During malignant diseases progression, changes in both DNA methylation patterns and DNA methyltransferase (MTase) genes are observed. Human de novo MTase DNMT3A is most frequently mutated in acute myeloid leukemia (AML) with a striking prevalence of R882H mutation, which has been extensively studied. Here, we investigate the functional role of the missense mutations (S714C, R635W, R736H, R771L, P777R, and F752V) found in the catalytic domain of DNMT3A in AML patients. These were accordingly mutated in the murine Dnmt3a catalytic domain (S124C, R45W, R146H, R181L, P187R, and F162V) and in addition, one-site CpG-containing DNA substrates were used as a model system. The 3-15-fold decrease (S124C and P187R) or complete loss (F162V, R45W, and R146H) of Dnmt3a-CD methylation activity was observed. Remarkably, Pro 187 and Arg 146 are not located at or near the Dnmt3a functional motives. Regulatory protein Dnmt3L did not enhance the methylation activity of R45W, R146H, P187R, and F162V mutants. The key steps of the Dnmt3a-mediated methylation mechanism, including DNA binding and transient covalent intermediate formation, were examined. There was a complete loss of DNA-binding affinity for R45W located in the AdoMet binding region and for R146H. Dnmt3a mutants studied in vitro suggest functional impairment of DNMT3A during pathogenesis.
Keywords: DNA methylation; DNA methyltransferase Dnmt3a; DNA-protein binding; S-adenosyl-L-methionine; leukemia; missense mutations.
Conflict of interest statement
Authors declare no conflict of interest in financial or any other sphere.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

