The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology
- PMID: 31861540
- PMCID: PMC7168646
- DOI: 10.3390/pathogens9010006
The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology
Abstract
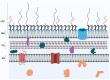
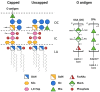
The major constituent of the outer membrane of Gram-negative bacteria is lipopolysaccharide (LPS), which is comprised of lipid A, core oligosaccharide, and O antigen, which is a long polysaccharide chain extending into the extracellular environment. Due to the localization of LPS, it is a key molecule on the bacterial cell wall that is recognized by the host to deploy an immune defence in order to neutralize invading pathogens. However, LPS also promotes bacterial survival in a host environment by protecting the bacteria from these threats. This review explores the relationship between the different LPS glycoforms of the opportunistic pathogen Pseudomonas aeruginosa and the ability of this organism to cause persistent infections, especially in the genetic disease cystic fibrosis. We also discuss the role of LPS in facilitating biofilm formation, antibiotic resistance, and how LPS may be targeted by new antimicrobial therapies.
Keywords: O antigen; antimicrobial resistance; biofilms; cystic fibrosis; host–pathogen interactions; lipopolysaccharide; pyocin.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

