Proteomic Analysis of Quercetin-Treated K562 Cells
- PMID: 31861640
- PMCID: PMC6981597
- DOI: 10.3390/ijms21010032
Proteomic Analysis of Quercetin-Treated K562 Cells
Abstract
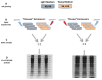
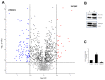
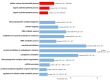
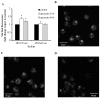
Among natural products under investigation for their additive potential in cancer prevention and treatment, the flavonoid quercetin has received attention for its effects on the cell cycle arrest and apoptosis. In the past, we addressed this issue in K562 cells, a cellular model of the human chronic myeloid leukemia. Here, we applied stable isotope labeling by amino acids in cell culture (SILAC) proteomics with the aim to increase knowledge on the regulative and metabolic pathways modulated by quercetin in these cells. After 24 h of quercetin treatment, we observed that apoptosis was not completely established, thus we selected this time range to capture quantitative data. As a result, we were able to achieve a robust identification of 1703 proteins, and to measure fold changes between quercetin-treated and untreated cells for 1206 proteins. Through a bioinformatics functional analysis on a subset of 112 proteins, we propose that the apoptotic phenotype of K562 cells entails a significant modulation of the translational machinery, RNA metabolism, antioxidant defense systems, and enzymes involved in lipid metabolism. Finally, we selected eight differentially expressed proteins, validated their modulated expression in quercetin-treated K562 cells, and discussed their possible role in flavonoid cytotoxicity. This quantitative profiling, performed for the first time on this type of tumor cells upon treatment with a flavonoid, will contribute to revealing the molecular basis of the multiplicity of the effects selectively exerted by quercetin on K562 cells.
Keywords: K562; SILAC; apoptosis; chronic myeloid leukemia; flavonoids; lipid metabolism; oxidative stress; quantitative proteomics; quercetin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

