Transcriptome Changes Induced by Different Potassium Levels in Banana Roots
- PMID: 31861661
- PMCID: PMC7020221
- DOI: 10.3390/plants9010011
Transcriptome Changes Induced by Different Potassium Levels in Banana Roots
Abstract
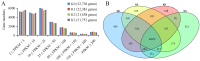
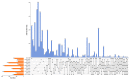
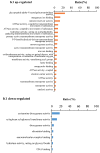
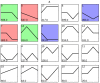
Potassium plays an important role in enhancing plant resistance to biological and abiotic stresses and improving fruit quality. To study the effect of potassium nutrient levels on banana root growth and its regulation mechanism, four potassium concentrations were designed to treat banana roots from no potassium to high potassium. The results indicated that K2 (3 mmol/L K2SO4) treatment was a relatively normal potassium concentration for the growth of banana root, and too high or too low potassium concentration was not conducive to the growth of banana root. By comparing the transcriptome data in each treatment in pairs, 4454 differentially expressed genes were obtained. There were obvious differences in gene function enrichment in root systems treated with different concentrations of potassium. Six significant expression profiles (profile 0, 1, 2, 7, 9 and 13) were identified by STEM analysis. The hub genes were FKF1, HsP70-1, NRT1/PTR5, CRY1, and ZIP11 in the profile 0; CYP51 in profile 1; SOS1 in profile 7; THA, LKR/SDH, MCC, C4H, CHI, F3'H, 2 PR1s, BSP, TLP, ICS, RO, chitinase and peroxidase in profile 9. Our results provide a comprehensive and systematic analysis of the gene regulation network in banana roots under different potassium stress.
Keywords: banana; potassium; root; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Genome-wide transcriptome analysis and identification of benzothiadiazole-induced genes and pathways potentially associated with defense response in banana.BMC Genomics. 2018 Jun 13;19(1):454. doi: 10.1186/s12864-018-4830-7. BMC Genomics. 2018. PMID: 29898655 Free PMC article.
-
Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense.BMC Genomics. 2013 Dec 5;14(1):851. doi: 10.1186/1471-2164-14-851. BMC Genomics. 2013. PMID: 24304681 Free PMC article.
-
Transcriptome and Gene Co-Expression Network Analysis Identifying Differentially Expressed Genes and Signal Pathways Involved in the Height Development of Banana (Musa spp.).Int J Mol Sci. 2023 Jan 30;24(3):2628. doi: 10.3390/ijms24032628. Int J Mol Sci. 2023. PMID: 36768952 Free PMC article.
-
Effects of Mycorrhizal Colonization on Transcriptional Expression of the Responsive Factor JERF3 and Stress-Responsive Genes in Banana Plantlets in Response to Combined Biotic and Abiotic Stresses.Front Plant Sci. 2021 Oct 28;12:742628. doi: 10.3389/fpls.2021.742628. eCollection 2021. Front Plant Sci. 2021. PMID: 34777424 Free PMC article.
-
Transcriptome analysis and identification of the low potassium stress-responsive gene SiSnRK2.6 in foxtail millet (Setaria italica L.).Theor Appl Genet. 2024 Jan 16;137(1):22. doi: 10.1007/s00122-023-04532-6. Theor Appl Genet. 2024. PMID: 38227064 Review.
Cited by
-
Omics-driven crop potassium use efficiency breeding.Front Plant Sci. 2022 Nov 24;13:1076193. doi: 10.3389/fpls.2022.1076193. eCollection 2022. Front Plant Sci. 2022. PMID: 36507409 Free PMC article. No abstract available.
-
Transcriptome Analysis of Pyrus betulaefolia Seedling Root Responses to Short-Term Potassium Deficiency.Int J Mol Sci. 2020 Nov 23;21(22):8857. doi: 10.3390/ijms21228857. Int J Mol Sci. 2020. PMID: 33238495 Free PMC article.
-
Transcriptome Analysis Unravels Key Factors Involved in Response to Potassium Deficiency and Feedback Regulation of K+ Uptake in Cotton Roots.Int J Mol Sci. 2021 Mar 19;22(6):3133. doi: 10.3390/ijms22063133. Int J Mol Sci. 2021. PMID: 33808570 Free PMC article.
-
Genome-wide identification and characterization of filamentation temperature-sensitive H (FtsH) genes and expression analysis in response to multiple stresses in Medicago truncatula.Mol Biol Rep. 2023 Dec;50(12):10097-10109. doi: 10.1007/s11033-023-08851-1. Epub 2023 Nov 1. Mol Biol Rep. 2023. PMID: 37910387
-
Comparative Transcriptome Profiling of Cassava Tuberous Roots in Response to Postharvest Physiological Deterioration.Int J Mol Sci. 2022 Dec 23;24(1):246. doi: 10.3390/ijms24010246. Int J Mol Sci. 2022. PMID: 36613690 Free PMC article.
References
-
- Clarkson D.T., Hanson J.B. The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 2003;31:239–298. doi: 10.1146/annurev.pp.31.060180.001323. - DOI
-
- Leigh R.A., Jones R.G.W. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 1984;97:1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x. - DOI
-
- Ahanger M.A., Agarwal R.M., Tomar N.S., Shrivastava M. Potassium induces positive changes in nitrogen metabolism and antioxidant system of oat (Avena sativa L. cultivar Kent) J. Plant Interact. 2015;10:211–223. doi: 10.1080/17429145.2015.1056260. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

