Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells
- PMID: 31861681
- PMCID: PMC7017164
- DOI: 10.3390/cancers12010022
Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells
Abstract
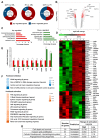
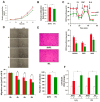
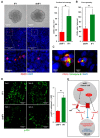
Increasing evidences show that the ATPase Inhibitory Factor 1 (IF1), the physiological inhibitor of the ATP synthase, is overexpressed in a large number of carcinomas contributing to metabolic reprogramming and cancer progression. Herein, we show that in contrast to the findings in other carcinomas, the overexpression of IF1 in a cohort of colorectal carcinomas (CRC) predicts less chances of disease recurrence, IF1 being an independent predictor of survival. Bioinformatic and gene expression analyses of the transcriptome of colon cancer cells with differential expression of IF1 indicate that cells overexpressing IF1 display a less aggressive behavior than IF1 silenced (shIF1) cells. Proteomic and functional in vitro migration and invasion assays confirmed the higher tumorigenic potential of shIF1 cells. Moreover, shIF1 cells have increased in vivo metastatic potential. The higher metastatic potential of shIF1 cells relies on increased cFLIP-mediated resistance to undergo anoikis after cell detachment. Furthermore, tumor spheroids of shIF1 cells have an increased ability to escape from immune surveillance by NK cells. Altogether, the results reveal that the overexpression of IF1 acts as a tumor suppressor in CRC with an important anti-metastatic role, thus supporting IF1 as a potential therapeutic target in CRC.
Keywords: ATPase Inhibitor Factor 1; colorectal cancer; immune surveillance; metastasis; prognosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Overexpression of the ATPase Inhibitory Factor 1 Favors a Non-metastatic Phenotype in Breast Cancer.Front Oncol. 2017 Apr 10;7:69. doi: 10.3389/fonc.2017.00069. eCollection 2017. Front Oncol. 2017. PMID: 28443245 Free PMC article.
-
The ATPase Inhibitory Factor 1 (IF1) Contributes to the Warburg Effect and Is Regulated by Its Phosphorylation in S39 by a Protein Kinase A-like Activity.Cancers (Basel). 2024 Feb 29;16(5):1014. doi: 10.3390/cancers16051014. Cancers (Basel). 2024. PMID: 38473373 Free PMC article. Review.
-
Expression, regulation and clinical relevance of the ATPase inhibitory factor 1 in human cancers.Oncogenesis. 2013 Apr 22;2(4):e46. doi: 10.1038/oncsis.2013.9. Oncogenesis. 2013. PMID: 23608753 Free PMC article.
-
A Review of the Inhibition of the Mitochondrial ATP Synthase by IF1 in vivo: Reprogramming Energy Metabolism and Inducing Mitohormesis.Front Physiol. 2018 Sep 19;9:1322. doi: 10.3389/fphys.2018.01322. eCollection 2018. Front Physiol. 2018. PMID: 30283362 Free PMC article. Review.
-
The Role of Mitochondrial H+-ATP Synthase in Cancer.Front Oncol. 2018 Mar 7;8:53. doi: 10.3389/fonc.2018.00053. eCollection 2018. Front Oncol. 2018. PMID: 29564224 Free PMC article. Review.
Cited by
-
Long Non-Coding RNA and microRNA Interplay in Colorectal Cancer and Their Effect on the Tumor Microenvironment.Cancers (Basel). 2022 Nov 5;14(21):5450. doi: 10.3390/cancers14215450. Cancers (Basel). 2022. PMID: 36358867 Free PMC article. Review.
-
Generation of mitochondrial reactive oxygen species is controlled by ATPase inhibitory factor 1 and regulates cognition.PLoS Biol. 2021 May 13;19(5):e3001252. doi: 10.1371/journal.pbio.3001252. eCollection 2021 May. PLoS Biol. 2021. PMID: 33983919 Free PMC article.
-
Development and Validation of an Anoikis-Related Gene Signature for Prognostic Prediction in Cervical Cancer.Int J Gen Med. 2025 Jun 4;18:2861-2879. doi: 10.2147/IJGM.S508059. eCollection 2025. Int J Gen Med. 2025. PMID: 40492231 Free PMC article.
-
Increased Expression of Mitochondrial UQCRC1 in Pancreatic Cancer Impairs Antitumor Immunity of Natural Killer Cells via Elevating Extracellular ATP.Front Oncol. 2022 Jun 13;12:872017. doi: 10.3389/fonc.2022.872017. eCollection 2022. Front Oncol. 2022. PMID: 35769718 Free PMC article.
-
Analysis of the metabolic proteome of lung adenocarcinomas by reverse-phase protein arrays (RPPA) emphasizes mitochondria as targets for therapy.Oncogenesis. 2022 May 9;11(1):24. doi: 10.1038/s41389-022-00400-y. Oncogenesis. 2022. PMID: 35534478 Free PMC article.

