Comparative Analysis and Expression Patterns of the PLP_deC Genes in Dendrobium officinale
- PMID: 31861760
- PMCID: PMC6981363
- DOI: 10.3390/ijms21010054
Comparative Analysis and Expression Patterns of the PLP_deC Genes in Dendrobium officinale
Abstract
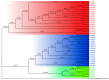
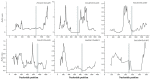
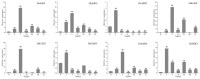
Studies have shown that the type II pyridoxal phosphate-dependent decarboxylase (PLP_deC) genes produce secondary metabolites and flavor volatiles in plants, and TDC (tryptophan decarboxylase), a member of the PLP_deC family, plays an important role in the biosynthesis of terpenoid indole alkaloids (TIAs). In this study, we identified eight PLP_deC genes in Dendrobium officinale (D. officinale) and six in Phalaenopsis equestris (P. equestris), and their structures, physicochemical properties, response elements, evolutionary relationships, and expression patterns were preliminarily predicted and analyzed. The results showed that PLP_deC genes play important roles in D. officinale and respond to different exogenous hormone treatments; additionally, the results support the selection of appropriate candidates for further functional characterization of PLP_deC genes in D. officinale.
Keywords: Dendrobium officinale; PLP_deC; bioinformatics; evolution; expression pattern analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Functional differentiation of olive PLP_deC genes: insights into metabolite biosynthesis and genetic improvement at the whole-genome level.Plant Cell Rep. 2024 Apr 23;43(5):127. doi: 10.1007/s00299-024-03212-z. Plant Cell Rep. 2024. PMID: 38652203
-
Characterization of LEA genes in Dendrobium officinale and one Gene in induction of callus.J Plant Physiol. 2021 Mar-Apr;258-259:153356. doi: 10.1016/j.jplph.2020.153356. Epub 2020 Dec 30. J Plant Physiol. 2021. PMID: 33423816
-
Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale.BMC Plant Biol. 2019 Jun 10;19(1):245. doi: 10.1186/s12870-019-1851-6. BMC Plant Biol. 2019. PMID: 31182022 Free PMC article.
-
Evolutionary Trails of Plant Group II Pyridoxal Phosphate-Dependent Decarboxylase Genes.Front Plant Sci. 2016 Aug 23;7:1268. doi: 10.3389/fpls.2016.01268. eCollection 2016. Front Plant Sci. 2016. PMID: 27602045 Free PMC article. Review.
-
Recent Advances and New Insights in Genome Analysis and Transcriptomic Approaches to Reveal Enzymes Associated with the Biosynthesis of Dendrobine-Type Sesquiterpenoid Alkaloids (DTSAs) from the Last Decade.Molecules. 2024 Aug 10;29(16):3787. doi: 10.3390/molecules29163787. Molecules. 2024. PMID: 39202866 Free PMC article. Review.
Cited by
-
Orchid Biochemistry.Int J Mol Sci. 2020 Mar 27;21(7):2338. doi: 10.3390/ijms21072338. Int J Mol Sci. 2020. PMID: 32230974 Free PMC article.
-
Natural Composition and Biosynthetic Pathways of Alkaloids in Medicinal Dendrobium Species.Front Plant Sci. 2022 May 6;13:850949. doi: 10.3389/fpls.2022.850949. eCollection 2022. Front Plant Sci. 2022. PMID: 35599884 Free PMC article. Review.
-
Micro-Evolution Analysis Reveals Diverged Patterns of Polyol Transporters in Seven Gramineae Crops.Front Genet. 2020 Jun 19;11:565. doi: 10.3389/fgene.2020.00565. eCollection 2020. Front Genet. 2020. PMID: 32636871 Free PMC article.
-
Comparison of the Antioxidant Activities and Polysaccharide Characterization of Fresh and Dry Dendrobium officinale Kimura et Migo.Molecules. 2022 Oct 7;27(19):6654. doi: 10.3390/molecules27196654. Molecules. 2022. PMID: 36235191 Free PMC article.
References
-
- Zhang J., He C., Wu K., Teixeira da Silva J.A., Zeng S., Zhang X., Yu Z., Xia H., Duan J. Transcriptome Analysis of Dendrobium officinale and its Application to the Identification of Genes Associated with Polysaccharide Synthesis. Front. Plant Sci. 2016;7:5. doi: 10.3389/fpls.2016.00005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

