Genome Analyses and Genome-Centered Metatranscriptomics of Methanothermobacter wolfeii Strain SIV6, Isolated from a Thermophilic Production-Scale Biogas Fermenter
- PMID: 31861790
- PMCID: PMC7022856
- DOI: 10.3390/microorganisms8010013
Genome Analyses and Genome-Centered Metatranscriptomics of Methanothermobacter wolfeii Strain SIV6, Isolated from a Thermophilic Production-Scale Biogas Fermenter
Abstract
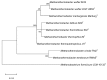
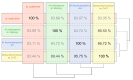
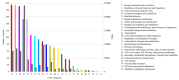
In the thermophilic biogas-producing microbial community, the genus Methanothermobacter was previously described to be frequently abundant. The aim of this study was to establish and analyze the genome sequence of the archaeal strain Methanothermobacter wolfeii SIV6 originating from a thermophilic industrial-scale biogas fermenter and compare it to related reference genomes. The circular chromosome has a size of 1,686,891 bases, featuring a GC content of 48.89%. Comparative analyses considering three completely sequenced Methanothermobacter strains revealed a core genome of 1494 coding sequences and 16 strain specific genes for M. wolfeii SIV6, which include glycosyltransferases and CRISPR/cas associated genes. Moreover, M. wolfeii SIV6 harbors all genes for the hydrogenotrophic methanogenesis pathway and genome-centered metatranscriptomics indicates the high metabolic activity of this strain, with 25.18% of all transcripts per million (TPM) belong to the hydrogenotrophic methanogenesis pathway and 18.02% of these TPM exclusively belonging to the mcr operon. This operon encodes the different subunits of the enzyme methyl-coenzyme M reductase (EC: 2.8.4.1), which catalyzes the final and rate-limiting step during methanogenesis. Finally, fragment recruitment of metagenomic reads from the thermophilic biogas fermenter on the SIV6 genome showed that the strain is abundant (1.2%) within the indigenous microbial community. Detailed analysis of the archaeal isolate M. wolfeii SIV6 indicates its role and function within the microbial community of the thermophilic biogas fermenter, towards a better understanding of the biogas production process and a microbial-based management of this complex process.
Keywords: CRISPR/cas; Methanothermobacter wolfeii; comparative analyses; fragment recruitment; genome mining; metabolic pathway reconstruction; metagenomics; metatranscriptomics; thermophilic biogas fermenter.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Complete Genome Sequence of Methanothermobacter sp. Strain THM-1, a Thermophilic and Hydrogenotrophic Methanogen Isolated from an Anaerobic Reactor in South Korea.Microbiol Resour Announc. 2021 Sep 23;10(38):e0058721. doi: 10.1128/MRA.00587-21. Epub 2021 Sep 23. Microbiol Resour Announc. 2021. PMID: 34553993 Free PMC article.
-
Phenotypic and genomic characterization of Methanothermobacter wolfeii strain BSEL, a CO2-capturing archaeon with minimal nutrient requirements.Appl Environ Microbiol. 2024 May 21;90(5):e0026824. doi: 10.1128/aem.00268-24. Epub 2024 Apr 15. Appl Environ Microbiol. 2024. PMID: 38619268 Free PMC article.
-
Genomic characterization of Defluviitoga tunisiensis L3, a key hydrolytic bacterium in a thermophilic biogas plant and its abundance as determined by metagenome fragment recruitment.J Biotechnol. 2016 Aug 20;232:50-60. doi: 10.1016/j.jbiotec.2016.05.001. Epub 2016 May 7. J Biotechnol. 2016. PMID: 27165504
-
The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology.J Biotechnol. 2008 Aug 31;136(1-2):77-90. doi: 10.1016/j.jbiotec.2008.05.008. Epub 2008 May 27. J Biotechnol. 2008. PMID: 18597880
-
An overview of physico-chemical mechanisms of biogas production by microbial communities: a step towards sustainable waste management.3 Biotech. 2016 Jun;6(1):72. doi: 10.1007/s13205-016-0395-9. Epub 2016 Feb 16. 3 Biotech. 2016. PMID: 28330142 Free PMC article. Review.
Cited by
-
Core cooperative metabolism in low-complexity CO2-fixing anaerobic microbiota.ISME J. 2025 Jan 2;19(1):wraf017. doi: 10.1093/ismejo/wraf017. ISME J. 2025. PMID: 39893570 Free PMC article.
-
Complete Genome Sequence of Methanothermobacter sp. Strain THM-1, a Thermophilic and Hydrogenotrophic Methanogen Isolated from an Anaerobic Reactor in South Korea.Microbiol Resour Announc. 2021 Sep 23;10(38):e0058721. doi: 10.1128/MRA.00587-21. Epub 2021 Sep 23. Microbiol Resour Announc. 2021. PMID: 34553993 Free PMC article.
-
Uncovering Microbiome Adaptations in a Full-Scale Biogas Plant: Insights from MAG-Centric Metagenomics and Metaproteomics.Microorganisms. 2023 Sep 27;11(10):2412. doi: 10.3390/microorganisms11102412. Microorganisms. 2023. PMID: 37894070 Free PMC article.
-
The novel oligopeptide utilizing species Anaeropeptidivorans aminofermentans M3/9T, its role in anaerobic digestion and occurrence as deduced from large-scale fragment recruitment analyses.Front Microbiol. 2022 Nov 9;13:1032515. doi: 10.3389/fmicb.2022.1032515. eCollection 2022. Front Microbiol. 2022. PMID: 36439843 Free PMC article.
-
Comparison of microbiome community structure and dynamics during anaerobic digestion of different renewable solid wastes.Curr Res Microb Sci. 2025 Mar 31;8:100383. doi: 10.1016/j.crmicr.2025.100383. eCollection 2025. Curr Res Microb Sci. 2025. PMID: 40255248 Free PMC article.
References
-
- Zinder S.H. Methanogenesis. Springer; Berlin/Heidelberg, Germany: 1993. Physiological ecology of methanogens; pp. 128–206.
-
- Conrad R. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 1999;28:193–202. doi: 10.1111/j.1574-6941.1999.tb00575.x. - DOI
-
- Stolze Y., Bremges A., Rumming M., Henke C., Maus I., Pühler A., Sczyrba A., Schlüter A. Identification and genome reconstruction of abundant distinct taxa in microbiomes from one thermophilic and three mesophilic production-scale biogas plants. Biotechnol. Biofuels. 2016;9:156. doi: 10.1186/s13068-016-0565-3. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

