A unified STR profiling system across multiple species with whole genome sequencing data
- PMID: 31861983
- PMCID: PMC6923897
- DOI: 10.1186/s12859-019-3246-y
A unified STR profiling system across multiple species with whole genome sequencing data
Abstract
Background: Short tandem repeats (STRs) serve as genetic markers in forensic scenes due to their high polymorphism in eukaryotic genomes. A variety of STRs profiling systems have been developed for species including human, dog, cat, cattle, etc. Maintaining these systems simultaneously can be costly. These mammals share many high similar regions along their genomes. With the availability of the massive amount of the whole genomics data of these species, it is possible to develop a unified STR profiling system. In this study, our objective is to propose and develop a unified set of STR loci that could be simultaneously applied to multiple species.
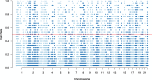
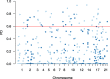
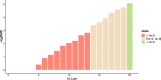
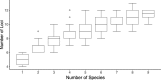
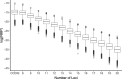
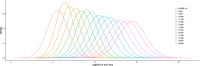
Result: To find a unified STR set, we collected the whole genome sequence data of the concerned species and mapped them to the human genome reference. Then we extracted the STR loci across the species. From these loci, we proposed an algorithm which selected a subset of loci by incorporating the optimized combined power of discrimination. Our results show that the unified set of loci have high combined power of discrimination, >1-10-9, for both individual species and the mixed population, as well as the random-match probability, <10-7 for all the involved species, indicating that the identified set of STR loci could be applied to multiple species.
Conclusions: We identified a set of STR loci which shared by multiple species. It implies that a unified STR profiling system is possible for these species under the forensic scenes. The system can be applied to the individual identification or paternal test of each of the ten common species which are Sus scrofa (pig), Bos taurus (cattle), Capra hircus (goat), Equus caballus (horse), Canis lupus familiaris (dog), Felis catus (cat), Ovis aries (sheep), Oryctolagus cuniculus (rabbit), and Bos grunniens (yak), and Homo sapiens (human). Our loci selection algorithm employed a greedy approach. The algorithm can generate the loci under different forensic parameters and for a specific combination of species.
Keywords: Individual identification; Short tandem repeats; Whole genome sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Development and Validation of a Novel Five-Dye Short Tandem Repeat Panel for Forensic Identification of 11 Species.Front Genet. 2020 Sep 24;11:1005. doi: 10.3389/fgene.2020.01005. eCollection 2020. Front Genet. 2020. PMID: 33193588 Free PMC article.
-
STRScan: targeted profiling of short tandem repeats in whole-genome sequencing data.BMC Bioinformatics. 2017 Oct 3;18(Suppl 11):398. doi: 10.1186/s12859-017-1800-z. BMC Bioinformatics. 2017. PMID: 28984185 Free PMC article.
-
Search for More Effective Microsatellite Markers for Forensics With Next-Generation Sequencing.IEEE Trans Nanobioscience. 2017 Jul;16(5):375-381. doi: 10.1109/TNB.2017.2712795. Epub 2017 Jun 7. IEEE Trans Nanobioscience. 2017. PMID: 28600257
-
Biology and Genetics of New Autosomal STR Loci Useful for Forensic DNA Analysis.Forensic Sci Rev. 2012 Jan;24(1):15-26. Forensic Sci Rev. 2012. PMID: 26231356 Review.
-
"New turns from old STaRs": enhancing the capabilities of forensic short tandem repeat analysis.Electrophoresis. 2014 Nov;35(21-22):3173-87. doi: 10.1002/elps.201400095. Epub 2014 Jul 16. Electrophoresis. 2014. PMID: 24888494 Review.
Cited by
-
Application of Next-Generation Sequencing (NGS) Techniques for Selected Companion Animals.Animals (Basel). 2024 May 27;14(11):1578. doi: 10.3390/ani14111578. Animals (Basel). 2024. PMID: 38891625 Free PMC article. Review.
-
The International Conference on Intelligent Biology and Medicine (ICIBM) 2019: bioinformatics methods and applications for human diseases.BMC Bioinformatics. 2019 Dec 20;20(Suppl 24):676. doi: 10.1186/s12859-019-3240-4. BMC Bioinformatics. 2019. PMID: 31861973 Free PMC article.
-
Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping.Life (Basel). 2025 Apr 17;15(4):664. doi: 10.3390/life15040664. Life (Basel). 2025. PMID: 40283218 Free PMC article.
-
Development and Validation of a Novel Five-Dye Short Tandem Repeat Panel for Forensic Identification of 11 Species.Front Genet. 2020 Sep 24;11:1005. doi: 10.3389/fgene.2020.01005. eCollection 2020. Front Genet. 2020. PMID: 33193588 Free PMC article.
-
Origin, Evolution, and Research Development of Donkeys.Genes (Basel). 2022 Oct 25;13(11):1945. doi: 10.3390/genes13111945. Genes (Basel). 2022. PMID: 36360182 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

