Relaxed purifying selection in autopolyploids drives transposable element over-accumulation which provides variants for local adaptation
- PMID: 31862875
- PMCID: PMC6925279
- DOI: 10.1038/s41467-019-13730-0
Relaxed purifying selection in autopolyploids drives transposable element over-accumulation which provides variants for local adaptation
Abstract
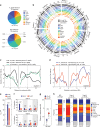
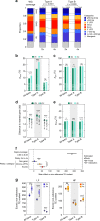
Polyploidization is frequently associated with increased transposable element (TE) content. However, what drives TE dynamics following whole genome duplication (WGD) and the evolutionary implications remain unclear. Here, we leverage whole-genome resequencing data available for ~300 individuals of Arabidopsis arenosa, a well characterized natural diploid-autotetraploid plant species, to address these questions. Based on 43,176 TE insertions we detect in these genomes, we demonstrate that relaxed purifying selection rather than transposition bursts is the main driver of TE over-accumulation after WGD. Furthermore, the increased pool of TE insertions in tetraploids is especially enriched within or near environmentally responsive genes. Notably, we show that the major flowering-time repressor gene FLC is disrupted by a TE insertion specifically in the rapid-cycling tetraploid lineage that colonized mainland railways. Together, our findings indicate that tetrasomy leads to an enhanced accumulation of genic TE insertions, some of which likely contribute to local adaptation.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- GREGORY T. RYAN. The Evolution of the Genome. 2005. Genome Size Evolution in Animals; pp. 3–87.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

