Mitochondrial oxidative capacity and NAD+ biosynthesis are reduced in human sarcopenia across ethnicities
- PMID: 31862890
- PMCID: PMC6925228
- DOI: 10.1038/s41467-019-13694-1
Mitochondrial oxidative capacity and NAD+ biosynthesis are reduced in human sarcopenia across ethnicities
Abstract
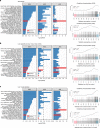
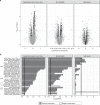
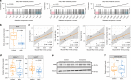
The causes of impaired skeletal muscle mass and strength during aging are well-studied in healthy populations. Less is known on pathological age-related muscle wasting and weakness termed sarcopenia, which directly impacts physical autonomy and survival. Here, we compare genome-wide transcriptional changes of sarcopenia versus age-matched controls in muscle biopsies from 119 older men from Singapore, Hertfordshire UK and Jamaica. Individuals with sarcopenia reproducibly demonstrate a prominent transcriptional signature of mitochondrial bioenergetic dysfunction in skeletal muscle, with low PGC-1α/ERRα signalling, and downregulation of oxidative phosphorylation and mitochondrial proteostasis genes. These changes translate functionally into fewer mitochondria, reduced mitochondrial respiratory complex expression and activity, and low NAD+ levels through perturbed NAD+ biosynthesis and salvage in sarcopenic muscle. We provide an integrated molecular profile of human sarcopenia across ethnicities, demonstrating a fundamental role of altered mitochondrial metabolism in the pathological loss of skeletal muscle mass and function in older people.
Conflict of interest statement
E.M., T.S., G.C., A.C., M.P.G., S.K., S. Mo, S. Me, J.M., F.R., P.D., and J.N.F. are full-time employees of the Société des Produits Nestlé (SPN). SA. K.M.G. and H.P.P. have received reimbursement for speaking at conferences sponsored by companies selling nutritional products. C.C. has received consultancy fees and honoraria from Amgen, Danone, Eli Lilly, GlaxoSmithKline, Medtronic, Merck, Nestlé, Novartis, Pfizer, Roche, Servier, Shire, Takeda, and UCB. K.M.G., Y.S.C., S.J.B., and N.K. are part of academic research programs that have received research funding from Abbott Nutrition, Nestec, and Danone. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

