Evolution of satellite plasmids can prolong the maintenance of newly acquired accessory genes in bacteria
- PMID: 31863068
- PMCID: PMC6925257
- DOI: 10.1038/s41467-019-13709-x
Evolution of satellite plasmids can prolong the maintenance of newly acquired accessory genes in bacteria
Abstract
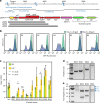
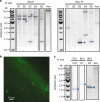
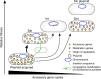
Transmissible plasmids spread genes encoding antibiotic resistance and other traits to new bacterial species. Here we report that laboratory populations of Escherichia coli with a newly acquired IncQ plasmid often evolve 'satellite plasmids' with deletions of accessory genes and genes required for plasmid replication. Satellite plasmids are molecular parasites: their presence reduces the copy number of the full-length plasmid on which they rely for their continued replication. Cells with satellite plasmids gain an immediate fitness advantage from reducing burdensome expression of accessory genes. Yet, they maintain copies of these genes and the complete plasmid, which potentially enables them to benefit from and transmit the traits they encode in the future. Evolution of satellite plasmids is transient. Cells that entirely lose accessory gene function or plasmid mobility dominate in the long run. Satellite plasmids also evolve in Snodgrassella alvi colonizing the honey bee gut, suggesting that this mechanism may broadly contribute to the importance of IncQ plasmids as agents of bacterial gene transfer in nature.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Diversification of Type VI Secretion System Toxins Reveals Ancient Antagonism among Bee Gut Microbes.mBio. 2017 Dec 12;8(6):e01630-17. doi: 10.1128/mBio.01630-17. mBio. 2017. PMID: 29233893 Free PMC article.
-
Geographical resistome profiling in the honeybee microbiome reveals resistance gene transfer conferred by mobilizable plasmids.Microbiome. 2022 May 3;10(1):69. doi: 10.1186/s40168-022-01268-1. Microbiome. 2022. PMID: 35501925 Free PMC article.
-
Glyphosate perturbs the gut microbiota of honey bees.Proc Natl Acad Sci U S A. 2018 Oct 9;115(41):10305-10310. doi: 10.1073/pnas.1803880115. Epub 2018 Sep 24. Proc Natl Acad Sci U S A. 2018. PMID: 30249635 Free PMC article.
-
Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context.Trends Microbiol. 2018 Dec;26(12):978-985. doi: 10.1016/j.tim.2018.06.007. Epub 2018 Jul 23. Trends Microbiol. 2018. PMID: 30049587 Review.
-
Impact of plasmid interactions with the chromosome and other plasmids on the spread of antibiotic resistance.Plasmid. 2018 Sep;99:82-88. doi: 10.1016/j.plasmid.2018.09.009. Epub 2018 Sep 19. Plasmid. 2018. PMID: 30240700 Review.
Cited by
-
The future of self-selecting and stable fermentations.J Ind Microbiol Biotechnol. 2020 Nov;47(11):993-1004. doi: 10.1007/s10295-020-02325-0. Epub 2020 Nov 2. J Ind Microbiol Biotechnol. 2020. PMID: 33136197 Free PMC article. Review.
-
Plasmid-driven strategies for clone success in Escherichia coli.Nat Commun. 2025 Apr 3;16(1):2921. doi: 10.1038/s41467-025-57940-1. Nat Commun. 2025. PMID: 40180894 Free PMC article.
-
Comparative genome analysis of the first Listeria monocytogenes core genome multi-locus sequence types CT2050 AND CT2051 strains with their close relatives.AIMS Microbiol. 2022 Mar 21;8(1):61-72. doi: 10.3934/microbiol.2022006. eCollection 2022. AIMS Microbiol. 2022. PMID: 35496987 Free PMC article. Review.
-
Deciphering the Structural Diversity and Classification of the Mobile Tigecycline Resistance Gene tet(X)-Bearing Plasmidome among Bacteria.mSystems. 2020 Apr 28;5(2):e00134-20. doi: 10.1128/mSystems.00134-20. mSystems. 2020. PMID: 32345737 Free PMC article.
-
Genome-wide annotation and comparative analysis of miniature inverted-repeat transposable elements (MITEs) in six pear species.Planta. 2025 Jun 16;262(2):29. doi: 10.1007/s00425-025-04750-w. Planta. 2025. PMID: 40524055

