A workflow for generating multi-strain genome-scale metabolic models of prokaryotes
- PMID: 31863076
- PMCID: PMC7017905
- DOI: 10.1038/s41596-019-0254-3
A workflow for generating multi-strain genome-scale metabolic models of prokaryotes
Abstract
Genome-scale models (GEMs) of bacterial strains' metabolism have been formulated and used over the past 20 years. Recently, with the number of genome sequences exponentially increasing, multi-strain GEMs have proved valuable to define the properties of a species. Here, through four major stages, we extend the original Protocol used to generate a GEM for a single strain to enable multi-strain GEMs: (i) obtain or generate a high-quality model of a reference strain; (ii) compare the genome sequence between a reference strain and target strains to generate a homology matrix; (iii) generate draft strain-specific models from the homology matrix; and (iv) manually curate draft models. These multi-strain GEMs can be used to study pan-metabolic capabilities and strain-specific differences across a species, thus providing insights into its range of lifestyles. Unlike the original Protocol, this procedure is scalable and can be partly automated with the Supplementary Jupyter notebook Tutorial. This Protocol Extension joins the ranks of other comparable methods for generating models such as CarveMe and KBase. This extension of the original Protocol takes on the order of weeks to multiple months to complete depending on the availability of a suitable reference model.
Conflict of interest statement
Competing interests:
The authors declare no competing interests.
Figures

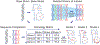

References
-
- Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev 2005; 15: 589–594. - PubMed
-
- Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Current Opinion in Microbiology. 2008; 11: 472–477. - PubMed
-
- Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet 2014; 15: 107–120. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

