CHC22 clathrin mediates traffic from early secretory compartments for human GLUT4 pathway biogenesis
- PMID: 31863584
- PMCID: PMC7039200
- DOI: 10.1083/jcb.201812135
CHC22 clathrin mediates traffic from early secretory compartments for human GLUT4 pathway biogenesis
Abstract
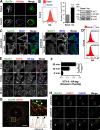
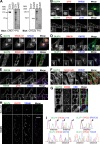
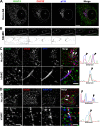
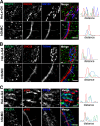
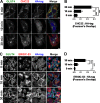
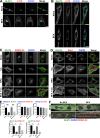
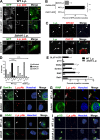
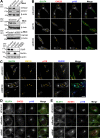
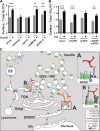
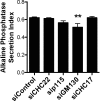
Glucose transporter 4 (GLUT4) is sequestered inside muscle and fat and then released by vesicle traffic to the cell surface in response to postprandial insulin for blood glucose clearance. Here, we map the biogenesis of this GLUT4 traffic pathway in humans, which involves clathrin isoform CHC22. We observe that GLUT4 transits through the early secretory pathway more slowly than the constitutively secreted GLUT1 transporter and localize CHC22 to the ER-to-Golgi intermediate compartment (ERGIC). CHC22 functions in transport from the ERGIC, as demonstrated by an essential role in forming the replication vacuole of Legionella pneumophila bacteria, which requires ERGIC-derived membrane. CHC22 complexes with ERGIC tether p115, GLUT4, and sortilin, and downregulation of either p115 or CHC22, but not GM130 or sortilin, abrogates insulin-responsive GLUT4 release. This indicates that CHC22 traffic initiates human GLUT4 sequestration from the ERGIC and defines a role for CHC22 in addition to retrograde sorting of GLUT4 after endocytic recapture, enhancing pathways for GLUT4 sequestration in humans relative to mice, which lack CHC22.
© 2019 Camus et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

