Native elongation transcript sequencing reveals temperature dependent dynamics of nascent RNAPII transcription in Arabidopsis
- PMID: 31863587
- PMCID: PMC7049701
- DOI: 10.1093/nar/gkz1189
Native elongation transcript sequencing reveals temperature dependent dynamics of nascent RNAPII transcription in Arabidopsis
Abstract
Temperature profoundly affects the kinetics of biochemical reactions, yet how large molecular complexes such as the transcription machinery accommodate changing temperatures to maintain cellular function is poorly understood. Here, we developed plant native elongating transcripts sequencing (plaNET-seq) to profile genome-wide nascent RNA polymerase II (RNAPII) transcription during the cold-response of Arabidopsis thaliana with single-nucleotide resolution. Combined with temporal resolution, these data revealed transient genome-wide reprogramming of nascent RNAPII transcription during cold, including characteristics of RNAPII elongation and thousands of non-coding transcripts connected to gene expression. Our results suggest a role for promoter-proximal RNAPII stalling in predisposing genes for transcriptional activation during plant-environment interactions. At gene 3'-ends, cold initially facilitated transcriptional termination by limiting the distance of read-through transcription. Within gene bodies, cold reduced the kinetics of co-transcriptional splicing leading to increased intragenic stalling. Our data resolved multiple distinct mechanisms by which temperature transiently altered the dynamics of nascent RNAPII transcription and associated RNA processing, illustrating potential biotechnological solutions and future focus areas to promote food security in the context of a changing climate.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
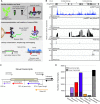
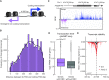
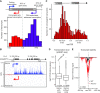
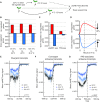


References
-
- Markovskaya E.F., Shibaeva T.G.. Low temperature sensors in plants: hypotheses and assumptions. Biol. Bull. 2017; 44:150–158.
-
- Thomashow M.F. PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999; 50:571–599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

