Unique repression domains of Pumilio utilize deadenylation and decapping factors to accelerate destruction of target mRNAs
- PMID: 31863588
- PMCID: PMC7038932
- DOI: 10.1093/nar/gkz1187
Unique repression domains of Pumilio utilize deadenylation and decapping factors to accelerate destruction of target mRNAs
Abstract
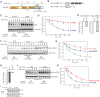
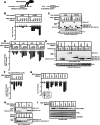
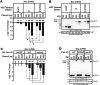
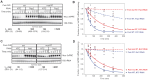
Pumilio is an RNA-binding protein that represses a network of mRNAs to control embryogenesis, stem cell fate, fertility and neurological functions in Drosophila. We sought to identify the mechanism of Pumilio-mediated repression and find that it accelerates degradation of target mRNAs, mediated by three N-terminal Repression Domains (RDs), which are unique to Pumilio orthologs. We show that the repressive activities of the Pumilio RDs depend on specific subunits of the Ccr4-Not (CNOT) deadenylase complex. Depletion of Pop2, Not1, Not2, or Not3 subunits alleviates Pumilio RD-mediated repression of protein expression and mRNA decay, whereas depletion of other CNOT components had little or no effect. Moreover, the catalytic activity of Pop2 deadenylase is important for Pumilio RD activity. Further, we show that the Pumilio RDs directly bind to the CNOT complex. We also report that the decapping enzyme, Dcp2, participates in repression by the N-terminus of Pumilio. These results support a model wherein Pumilio utilizes CNOT deadenylase and decapping complexes to accelerate destruction of target mRNAs. Because the N-terminal RDs are conserved in mammalian Pumilio orthologs, the results of this work broadly enhance our understanding of Pumilio function and roles in diseases including cancer, neurodegeneration and epilepsy.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







Similar articles
-
Human Pumilio proteins directly bind the CCR4-NOT deadenylase complex to regulate the transcriptome.RNA. 2021 Apr;27(4):445-464. doi: 10.1261/rna.078436.120. Epub 2021 Jan 4. RNA. 2021. PMID: 33397688 Free PMC article.
-
A conserved domain of Drosophila RNA-binding protein Pumilio interacts with multiple CCR4-NOT deadenylase complex subunits to repress target mRNAs.J Biol Chem. 2022 Sep;298(9):102270. doi: 10.1016/j.jbc.2022.102270. Epub 2022 Jul 16. J Biol Chem. 2022. PMID: 35850301 Free PMC article.
-
The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation.RNA. 2014 Aug;20(8):1298-319. doi: 10.1261/rna.046029.114. Epub 2014 Jun 18. RNA. 2014. PMID: 24942623 Free PMC article.
-
Post-transcriptional Regulatory Functions of Mammalian Pumilio Proteins.Trends Genet. 2018 Dec;34(12):972-990. doi: 10.1016/j.tig.2018.09.006. Epub 2018 Oct 10. Trends Genet. 2018. PMID: 30316580 Free PMC article. Review.
-
Eukaryotic mRNA decapping factors: molecular mechanisms and activity.FEBS J. 2023 Nov;290(21):5057-5085. doi: 10.1111/febs.16626. Epub 2022 Sep 30. FEBS J. 2023. PMID: 36098474 Free PMC article. Review.
Cited by
-
Functional significance of PUF partnerships in C. elegans germline stem cells.bioRxiv [Preprint]. 2023 Feb 16:2023.02.15.528708. doi: 10.1101/2023.02.15.528708. bioRxiv. 2023. Update in: Development. 2023 May 1;150(9):dev201705. doi: 10.1242/dev.201705. PMID: 36824876 Free PMC article. Updated. Preprint.
-
Human Pumilio proteins directly bind the CCR4-NOT deadenylase complex to regulate the transcriptome.RNA. 2021 Apr;27(4):445-464. doi: 10.1261/rna.078436.120. Epub 2021 Jan 4. RNA. 2021. PMID: 33397688 Free PMC article.
-
A higher order PUF complex is central to regulation of C. elegans germline stem cells.Nat Commun. 2025 Jan 2;16(1):123. doi: 10.1038/s41467-024-55526-x. Nat Commun. 2025. PMID: 39747099 Free PMC article.
-
Multivalent interactions with CCR4-NOT and PABPC1 determine mRNA repression efficiency by tristetraprolin.Nat Commun. 2025 Aug 13;16(1):7528. doi: 10.1038/s41467-025-62741-7. Nat Commun. 2025. PMID: 40804313 Free PMC article.
-
NORAD-Regulated Signaling Pathways in Breast Cancer Progression.Cancers (Basel). 2024 Feb 1;16(3):636. doi: 10.3390/cancers16030636. Cancers (Basel). 2024. PMID: 38339387 Free PMC article. Review.
References
-
- Wickens M., Bernstein D.S., Kimble J., Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002; 18:150–157. - PubMed
-
- Lehmann R., Nusslein-Volhard C. Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embyro. Nature. 1987; 329:167–170.
-
- Barker D.D., Wang C., Moore J., Dickinson L.K., Lehmann R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992; 6:2312–2326. - PubMed
-
- Murata Y., Wharton R.P. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995; 80:747–756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

