The sensor kinase BfmS controls production of outer membrane vesicles in Acinetobacter baumannii
- PMID: 31864291
- PMCID: PMC6925498
- DOI: 10.1186/s12866-019-1679-0
The sensor kinase BfmS controls production of outer membrane vesicles in Acinetobacter baumannii
Abstract
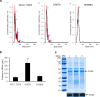
Background: Acinetobacter baumannii is an important opportunistic pathogen responsible for various nosocomial infections. The BfmRS two-component system plays a role in pathogenesis and antimicrobial resistance of A. baumannii via regulation of bacterial envelope structures. This study investigated the role of the sensor kinase, BfmS, in localization of outer membrane protein A (OmpA) in the outer membrane and production of outer membrane vesicles (OMVs) using wild-type A. baumannii ATCC 17978, ΔbfmS mutant, and bfmS-complemented strains.
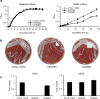
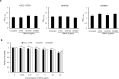
Results: The ΔbfmS mutant showed hypermucoid phenotype in the culture plates, growth retardation under static culture conditions, and reduced susceptibility to aztreonam and colistin compared to the wild-type strain. The ΔbfmS mutant produced less OmpA in the outer membrane but released more OmpA via OMVs than the wild-type strain, even though expression of ompA and its protein production were not different between the two strains. The ΔbfmS mutant produced 2.35 times more OMV particles and 4.46 times more OMV proteins than the wild-type stain. The ΔbfmS mutant OMVs were more cytotoxic towards A549 cells than wild-type strain OMVs.
Conclusions: The present study demonstrates that BfmS controls production of OMVs in A. baumannii. Moreover, BfmS negatively regulates antimicrobial resistance of A. baumannii and OMV-mediated host cell cytotoxicity. Our results indicate that BfmS negatively controls the pathogenic traits of A. baumannii via cell envelope structures and OMV production.
Keywords: Acinetobacter baumannii; BfmS; Cytotoxicity; OmpA; Outer membrane vesicle.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Martín-Aspas A, Guerrero-Sánchez FM, García-Colchero F, Rodríguez-Roca S, Girón-González JA. Differential characteristics of Acinetobacter baumannii colonization and infection: risk factors, clinical picture, and mortality. Infect Drug Resist. 2018;11:861–872. doi: 10.2147/IDR.S163944. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

