Structure of the Cardiac Sodium Channel
- PMID: 31866066
- PMCID: PMC6986426
- DOI: 10.1016/j.cell.2019.11.041
Structure of the Cardiac Sodium Channel
Abstract
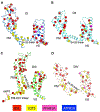
Voltage-gated sodium channel Nav1.5 generates cardiac action potentials and initiates the heartbeat. Here, we report structures of NaV1.5 at 3.2-3.5 Å resolution. NaV1.5 is distinguished from other sodium channels by a unique glycosyl moiety and loss of disulfide-bonding capability at the NaVβ subunit-interaction sites. The antiarrhythmic drug flecainide specifically targets the central cavity of the pore. The voltage sensors are partially activated, and the fast-inactivation gate is partially closed. Activation of the voltage sensor of Domain III allows binding of the isoleucine-phenylalanine-methionine (IFM) motif to the inactivation-gate receptor. Asp and Ala, in the selectivity motif DEKA, line the walls of the ion-selectivity filter, whereas Glu and Lys are in positions to accept and release Na+ ions via a charge-delocalization network. Arrhythmia mutation sites undergo large translocations during gating, providing a potential mechanism for pathogenic effects. Our results provide detailed insights into Nav1.5 structure, pharmacology, activation, inactivation, ion selectivity, and arrhythmias.
Keywords: antiarrhytymic drugs; cryoelectron microscopy arrhythmia; fast inactivation; gating pore current; heart; sodium channel; sodium selectivity.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests
The authors declare no competing interests.
Figures





Comment in
-
Say Cheese: Structure of the Cardiac Electrical Engine Is Captured.Trends Biochem Sci. 2020 May;45(5):369-371. doi: 10.1016/j.tibs.2020.02.003. Epub 2020 Feb 24. Trends Biochem Sci. 2020. PMID: 32311330
References
-
- Anno T, and Hondeghem LM (1990). Interactions of flecainide with guinea pig cardiac sodium channels. Importance of activation unblocking to the voltage dependence of recovery. Circ Res 66, 789–803. - PubMed
-
- Armstrong CM, and Bezanilla F (1973). Currents related to movement of the gating particles of the sodium channels. Nature 242, 459–461. - PubMed
-
- Baroudi G, Acharfi S, Larouche C, and Chahine M (2002). Expression and intracellular localization of an SCN5A double mutant R1232W/T1620M implicated in Brugada syndrome. Circ Res 90, E11–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

