Novel and Recurrent Mutations in a Cohort of Chinese Patients With Young-Onset Amyotrophic Lateral Sclerosis
- PMID: 31866807
- PMCID: PMC6908997
- DOI: 10.3389/fnins.2019.01289
Novel and Recurrent Mutations in a Cohort of Chinese Patients With Young-Onset Amyotrophic Lateral Sclerosis
Abstract
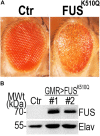
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that affects nerve cells in the brain and spinal cord. More than 25 ALS-related genes have been identified, accounting for approximately 10% of sporadic ALS (SALS) and two-thirds of familial ALS (FALS) cases. Several recent studies showed that genetic factors might have a larger contribution to young-onset ALS than to ALS cases overall. However, the genetic profile of young-onset ALS patients is not yet fully understood. Here, we investigated a cohort of 27 young-onset ALS patients (onset age < 45 years) through whole-exome sequencing (WES). Genetic analysis identified pathogenic variants of FUS (25.9%), SOD1 (22.2%), TARDBP (3.7%), and VCP (3.7%) in 27 young-onset ALS patients. Of 12 identified types of mutations, c.1528A > C in FUS and c.266G > A in VCP were novel. All of the cases in this study reflect a monogenic origin with an autosomal dominant mode of inheritance. Notably, a novel de novo missense mutation, c.1528A > C (p.K510Q), in FUS was identified in a 29-year-old ALS patient. Expression of the K510Q mutant FUS resulted in cytoplasmic mislocalization of FUS in cultured cells and induced neural toxicity in a fly model. This study provides further evidence of the genetic profile of young-onset ALS patients from China and expands the mutational spectrum of the FUS gene, with one new K510Q mutation identified.
Keywords: Drosophila model; amyotrophic lateral sclerosis; c.1528A > C (p. K510Q); fused in sarcoma; novel mutation; young-onset.
Copyright © 2019 Deng, Wu, Xie, Gang, Yu, Liu, Wang, Lv, Zhang, Huang, Wang, Yuan, Hong and Wang.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

