Coordinated Regulation of Rsd and RMF for Simultaneous Hibernation of Transcription Apparatus and Translation Machinery in Stationary-Phase Escherichia coli
- PMID: 31867037
- PMCID: PMC6904343
- DOI: 10.3389/fgene.2019.01153
Coordinated Regulation of Rsd and RMF for Simultaneous Hibernation of Transcription Apparatus and Translation Machinery in Stationary-Phase Escherichia coli
Abstract
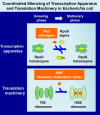
Transcription and translation in growing phase of Escherichia coli, the best-studied model prokaryote, are coupled and regulated in coordinate fashion. Accordingly, the growth rate-dependent control of the synthesis of RNA polymerase (RNAP) core enzyme (the core component of transcription apparatus) and ribosomes (the core component of translation machinery) is tightly coordinated to keep the relative level of transcription apparatus and translation machinery constant for effective and efficient utilization of resources and energy. Upon entry into the stationary phase, transcription apparatus is modulated by replacing RNAP core-associated sigma (promoter recognition subunit) from growth-related RpoD to stationary-phase-specific RpoS. The anti-sigma factor Rsd participates for the efficient replacement of sigma, and the unused RpoD is stored silent as Rsd-RpoD complex. On the other hand, functional 70S ribosome is transformed into inactive 100S dimer by two regulators, ribosome modulation factor (RMF) and hibernation promoting factor (HPF). In this review article, we overview how we found these factors and what we know about the molecular mechanisms for silencing transcription apparatus and translation machinery by these factors. In addition, we provide our recent findings of promoter-specific transcription factor (PS-TF) screening of the transcription factors involved in regulation of the rsd and rmf genes. Results altogether indicate the coordinated regulation of Rsd and RMF for simultaneous hibernation of transcription apparatus and translation machinery.
Keywords: Escherichia coli K-12; RNA polymerase sigma factor; anti-sigma factor (Rsd); hibernation; ribosome; ribosome modulation factor; stationary phase.
Copyright © 2019 Yoshida, Wada, Shimada, Maki and Ishihama.
Figures






Similar articles
-
Metal-Responsive Transcription Factors Co-Regulate Anti-Sigma Factor (Rsd) and Ribosome Dimerization Factor Expression.Int J Mol Sci. 2023 Mar 1;24(5):4717. doi: 10.3390/ijms24054717. Int J Mol Sci. 2023. PMID: 36902154 Free PMC article.
-
Coordinated Hibernation of Transcriptional and Translational Apparatus during Growth Transition of Escherichia coli to Stationary Phase.mSystems. 2018 Sep 11;3(5):e00057-18. doi: 10.1128/mSystems.00057-18. eCollection 2018 Sep-Oct. mSystems. 2018. PMID: 30225374 Free PMC article.
-
Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival.Genes Cells. 1999 Mar;4(3):135-43. doi: 10.1046/j.1365-2443.1999.00247.x. Genes Cells. 1999. PMID: 10320479 Review.
-
Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase.Genes Cells. 2009 Feb;14(2):271-80. doi: 10.1111/j.1365-2443.2008.01272.x. Epub 2008 Jan 15. Genes Cells. 2009. PMID: 19170772
-
Ribosome Hibernation.Annu Rev Genet. 2018 Nov 23;52:321-348. doi: 10.1146/annurev-genet-120215-035130. Annu Rev Genet. 2018. PMID: 30476446 Review.
Cited by
-
The Identity of the Constriction Region of the Ribosomal Exit Tunnel Is Important to Maintain Gene Expression in Escherichia coli.Microbiol Spectr. 2022 Apr 27;10(2):e0226121. doi: 10.1128/spectrum.02261-21. Epub 2022 Mar 21. Microbiol Spectr. 2022. PMID: 35311583 Free PMC article.
-
Ribosomal Hibernation Factor Links Quorum-Sensing to Acid Resistance in EHEC.Microorganisms. 2025 Jul 24;13(8):1730. doi: 10.3390/microorganisms13081730. Microorganisms. 2025. PMID: 40871233 Free PMC article.
-
Metal-Responsive Transcription Factors Co-Regulate Anti-Sigma Factor (Rsd) and Ribosome Dimerization Factor Expression.Int J Mol Sci. 2023 Mar 1;24(5):4717. doi: 10.3390/ijms24054717. Int J Mol Sci. 2023. PMID: 36902154 Free PMC article.
-
Genome-wide mapping of fluoroquinolone-stabilized DNA gyrase cleavage sites displays drug specific effects that correlate with bacterial persistence.Nucleic Acids Res. 2023 Feb 22;51(3):1208-1228. doi: 10.1093/nar/gkac1223. Nucleic Acids Res. 2023. PMID: 36631985 Free PMC article.
-
Pseudomonas aeruginosa Citrate Synthase GltA Influences Antibiotic Tolerance and the Type III Secretion System through the Stringent Response.Microbiol Spectr. 2023 Feb 14;11(1):e0323922. doi: 10.1128/spectrum.03239-22. Epub 2023 Jan 5. Microbiol Spectr. 2023. PMID: 36602339 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

