Towards Improved Molecular Identification Tools in Fine Fescue (Festuca L., Poaceae) Turfgrasses: Nuclear Genome Size, Ploidy, and Chloroplast Genome Sequencing
- PMID: 31867041
- PMCID: PMC6909427
- DOI: 10.3389/fgene.2019.01223
Towards Improved Molecular Identification Tools in Fine Fescue (Festuca L., Poaceae) Turfgrasses: Nuclear Genome Size, Ploidy, and Chloroplast Genome Sequencing
Abstract
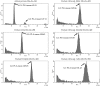
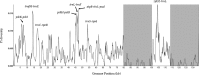
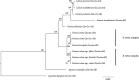
Fine fescues (Festuca L., Poaceae) are turfgrass species that perform well in low-input environments. Based on morphological characteristics, the most commonly-utilized fine fescues are divided into five taxa: three are subspecies within F. rubra L. and the remaining two are treated as species within the F. ovina L. complex. Morphologically, these five taxa are very similar; both identification and classification of fine fescues remain challenging. In an effort to develop identification methods for fescues, we used flow cytometry to estimate genome size and ploidy level and sequenced the chloroplast genome of all five taxa. Fine fescue chloroplast genome sizes ranged from 133,331 to 133,841 bp and contained 113-114 genes. Phylogenetic relationship reconstruction using whole chloroplast genome sequences agreed with previous work based on morphology. Comparative genomics suggested unique repeat signatures for each fine fescue taxon that could potentially be used for marker development for taxon identification.
Keywords: chloroplast genome; comparative genomics; fine fescue; low input turfgrass; phylogeny.
Copyright © 2019 Qiu, Hirsch, Yang and Watkins.
Figures





Similar articles
-
Molecular discrimination of tall fescue morphotypes in association with Festuca relatives.PLoS One. 2018 Jan 17;13(1):e0191343. doi: 10.1371/journal.pone.0191343. eCollection 2018. PLoS One. 2018. PMID: 29342197 Free PMC article.
-
First insight into the phylogeny of fine-leaved Festuca in the Altai Mountain Country based on genome-wide genotyping.Ecol Evol. 2023 Apr 2;13(4):e9943. doi: 10.1002/ece3.9943. eCollection 2023 Apr. Ecol Evol. 2023. PMID: 37021080 Free PMC article.
-
Low variability of internal transcribed spacer rDNA and trnL (UAA) intron sequences of several taxa in the Festuca ovina aggregate (POACEAE).Acta Biol Hung. 2006 Mar;57(1):57-69. doi: 10.1556/ABiol.57.2006.1.6. Acta Biol Hung. 2006. PMID: 16646525
-
Development and characterization of chloroplast microsatellite markers in a fine-leaved fescue, Festuca rubra (Poaceae).Appl Plant Sci. 2014 Dec 4;2(12):apps.1400094. doi: 10.3732/apps.1400094. eCollection 2014 Dec. Appl Plant Sci. 2014. PMID: 25506524 Free PMC article.
-
Breeding for disease resistance in the major cool-season turfgrasses.Annu Rev Phytopathol. 2006;44:213-34. doi: 10.1146/annurev.phyto.44.070505.143338. Annu Rev Phytopathol. 2006. PMID: 17061916 Review.
Cited by
-
Comparative and Phylogenetic Analysis of Complete Chloroplast Genomes in Leymus (Triticodae, Poaceae).Genes (Basel). 2022 Aug 10;13(8):1425. doi: 10.3390/genes13081425. Genes (Basel). 2022. PMID: 36011336 Free PMC article.
-
Analysis of Chloroplast Genomes Provides Insights Into the Evolution of Agropyron.Front Genet. 2022 Jan 25;13:832809. doi: 10.3389/fgene.2022.832809. eCollection 2022. Front Genet. 2022. PMID: 35145553 Free PMC article.
-
Characterization and Comparative Analysis of Complete Chloroplast Genomes of Four Bromus (Poaceae, Bromeae) Species.Genes (Basel). 2024 Jun 20;15(6):815. doi: 10.3390/genes15060815. Genes (Basel). 2024. PMID: 38927750 Free PMC article.
-
Comparative Plastomes and Phylogenetic Analysis of Cleistogenes and Closely Related Genera (Poaceae).Front Plant Sci. 2021 Mar 25;12:638597. doi: 10.3389/fpls.2021.638597. eCollection 2021. Front Plant Sci. 2021. PMID: 33841465 Free PMC article.
-
Comparative analysis of two Korean irises (Iris ruthenica and I. uniflora, Iridaceae) based on plastome sequencing and micromorphology.Sci Rep. 2022 Jun 8;12(1):9424. doi: 10.1038/s41598-022-13528-z. Sci Rep. 2022. PMID: 35676304 Free PMC article.
References
-
- Arumuganathan K., Tallury S., Fraser M., Bruneau A., Qu R. (1999). Nuclear DNA content of thirteen turfgrass species by flow cytometry. Crop Sci. 39, 1518–1521. 10.2135/cropsci1999.3951518x - DOI
-
- Baldwin B. G., Sanderson M. J., Porter J. M., Wojciechowski M. F., Campbell C. S., Donoghue M. J. (1995). The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Garden, 82 (2), 247–277. 10.2307/2399880 - DOI
-
- Beard J. B. (1972). Turfgrass: Science and culture.
-
- Bonos S. A., Huff D. R. (2013). Cool-season grasses: Biology and breeding. Turfgrass. Biology, Use Manage 7, 591–660. 10.2134/agronmonogr56.c17 - DOI
LinkOut - more resources
Full Text Sources

