ProDCoNN: Protein design using a convolutional neural network
- PMID: 31867753
- PMCID: PMC8204568
- DOI: 10.1002/prot.25868
ProDCoNN: Protein design using a convolutional neural network
Abstract
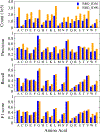
Designing protein sequences that fold to a given three-dimensional (3D) structure has long been a challenging problem in computational structural biology with significant theoretical and practical implications. In this study, we first formulated this problem as predicting the residue type given the 3D structural environment around the C α atom of a residue, which is repeated for each residue of a protein. We designed a nine-layer 3D deep convolutional neural network (CNN) that takes as input a gridded box with the atomic coordinates and types around a residue. Several CNN layers were designed to capture structure information at different scales, such as bond lengths, bond angles, torsion angles, and secondary structures. Trained on a very large number of protein structures, the method, called ProDCoNN (protein design with CNN), achieved state-of-the-art performance when tested on large numbers of test proteins and benchmark datasets.
Keywords: ProDCoNN; convolutional neural network; inverse folding problem; protein design; protein engineering.
© 2019 Wiley Periodicals, Inc.
Figures


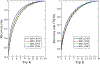



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

