Deep Residual Neural Networks Resolve Quartet Molecular Phylogenies
- PMID: 31868908
- PMCID: PMC8453599
- DOI: 10.1093/molbev/msz307
Deep Residual Neural Networks Resolve Quartet Molecular Phylogenies
Abstract
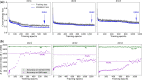
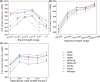
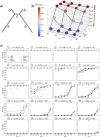
Phylogenetic inference is of fundamental importance to evolutionary as well as other fields of biology, and molecular sequences have emerged as the primary data for this task. Although many phylogenetic methods have been developed to explicitly take into account substitution models of sequence evolution, such methods could fail due to model misspecification or insufficiency, especially in the face of heterogeneities in substitution processes across sites and among lineages. In this study, we propose to infer topologies of four-taxon trees using deep residual neural networks, a machine learning approach needing no explicit modeling of the subject system and having a record of success in solving complex nonlinear inference problems. We train residual networks on simulated protein sequence data with extensive amino acid substitution heterogeneities. We show that the well-trained residual network predictors can outperform existing state-of-the-art inference methods such as the maximum likelihood method on diverse simulated test data, especially under extensive substitution heterogeneities. Reassuringly, residual network predictors generally agree with existing methods in the trees inferred from real phylogenetic data with known or widely believed topologies. Furthermore, when combined with the quartet puzzling algorithm, residual network predictors can be used to reconstruct trees with more than four taxa. We conclude that deep learning represents a powerful new approach to phylogenetic reconstruction, especially when sequences evolve via heterogeneous substitution processes. We present our best trained predictor in a freely available program named Phylogenetics by Deep Learning (PhyDL, https://gitlab.com/ztzou/phydl; last accessed January 3, 2020).
Keywords: deep learning; heterotachy; long-branch attraction; phylogenetic inference; protein sequence evolution; residual neural network.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Machine learning can be as good as maximum likelihood when reconstructing phylogenetic trees and determining the best evolutionary model on four taxon alignments.Mol Phylogenet Evol. 2024 Nov;200:108181. doi: 10.1016/j.ympev.2024.108181. Epub 2024 Aug 30. Mol Phylogenet Evol. 2024. PMID: 39209046
-
Reliable estimation of tree branch lengths using deep neural networks.PLoS Comput Biol. 2024 Aug 5;20(8):e1012337. doi: 10.1371/journal.pcbi.1012337. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39102450 Free PMC article.
-
Phyloformer: Fast, Accurate, and Versatile Phylogenetic Reconstruction with Deep Neural Networks.Mol Biol Evol. 2025 Apr 1;42(4):msaf051. doi: 10.1093/molbev/msaf051. Mol Biol Evol. 2025. PMID: 40066802 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
Applications of machine learning in phylogenetics.Mol Phylogenet Evol. 2024 Jul;196:108066. doi: 10.1016/j.ympev.2024.108066. Epub 2024 Mar 31. Mol Phylogenet Evol. 2024. PMID: 38565358 Review.
Cited by
-
phastSim: Efficient simulation of sequence evolution for pandemic-scale datasets.PLoS Comput Biol. 2022 Apr 29;18(4):e1010056. doi: 10.1371/journal.pcbi.1010056. eCollection 2022 Apr. PLoS Comput Biol. 2022. PMID: 35486906 Free PMC article.
-
A quartet-based approach for inferring phylogenetically informative features from genomic and phenomic data.Comput Struct Biotechnol J. 2025 Aug 22;27:3710-3718. doi: 10.1016/j.csbj.2025.08.015. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40895284 Free PMC article.
-
Exploring geometry of genome space via Grassmann manifolds.Innovation (Camb). 2024 Jul 22;5(5):100677. doi: 10.1016/j.xinn.2024.100677. eCollection 2024 Sep 9. Innovation (Camb). 2024. PMID: 39206218 Free PMC article.
-
DEPP: Deep Learning Enables Extending Species Trees using Single Genes.Syst Biol. 2023 May 19;72(1):17-34. doi: 10.1093/sysbio/syac031. Syst Biol. 2023. PMID: 35485976 Free PMC article.
-
Epi-DNNs: Epidemiological priors informed deep neural networks for modeling COVID-19 dynamics.Comput Biol Med. 2023 May;158:106693. doi: 10.1016/j.compbiomed.2023.106693. Epub 2023 Feb 28. Comput Biol Med. 2023. PMID: 36996662 Free PMC article.
References
-
- Atkinson QD, Meade A, Venditti C, Greenhill SJ, Pagel M. 2008. Languages evolve in punctuational bursts. Science 319(5863):588–588. - PubMed
-
- Bhattacharya S. 2014. Science in court: disease detectives. Nature 506(7489):424–426. - PubMed
-
- Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. 2012. Epistasis as the primary factor in molecular evolution. Nature 490(7421):535–538. - PubMed
-
- Byng J, Chase M, Christenhusz M, Fay M, Judd W, Mabberley D, Sennikov A, Soltis D, Soltis P, Stevens P, et al.. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: aPG IV. Bot J Linn Soc. 181:1–20.
-
- Carvalho SB, Velo-Antón G, Tarroso P, Portela AP, Barata M, Carranza S, Moritz C, Possingham HP. 2017. Spatial conservation prioritization of biodiversity spanning the evolutionary continuum. Nat Ecol Evol. 1(6):151.. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

