Transcriptomic signature of gut microbiome-contacting cells in colon of spontaneously hypertensive rats
- PMID: 31869283
- PMCID: PMC7099409
- DOI: 10.1152/physiolgenomics.00087.2019
Transcriptomic signature of gut microbiome-contacting cells in colon of spontaneously hypertensive rats
Abstract
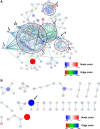
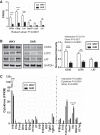
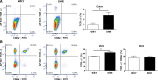
Fecal matter transfer from hypertensive patients and animals into normotensive animals increases blood pressure, strengthening the evidence for gut-microbiota interactions in the control of blood pressure. However, cellular and molecular events involved in gut dysbiosis-associated hypertension remain poorly understood. Therefore, our objective in this study was to use gene expression profiling to characterize the gut epithelium layer in the colon in hypertension. We observed significant suppression of components of T cell receptor (TCR) signaling in the colonic epithelium of spontaneously hypertensive rats (SHR) when compared with Wistar Kyoto (WKY) normotensive rats. Western blot analysis confirmed lower expression of key proteins including T cell surface glycoprotein CD3 gamma chain (Cd3g) and lymphocyte cytosolic protein 2 (Lcp2). Furthermore, lower expression of cytokines and receptors responsible for lymphocyte proliferation, differentiation, and activation (e.g., Il12r, Il15ra, Il7, Il16, Tgfb1) was observed in the colonic epithelium of the SHR. Finally, Alpi and its product, intestinal alkaline phosphatase, primarily localized in the epithelial cells, were profoundly lower in the SHR. These observations demonstrate that the colonic epithelium undergoes functional changes linked to altered immune, barrier function, and dysbiosis in hypertension.
Keywords: TCR signaling pathway; colonic epithelium; gene expression profile; hypertension.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

