Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations
- PMID: 31869403
- PMCID: PMC6953885
- DOI: 10.1371/journal.pgen.1008500
Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations
Abstract
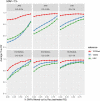
Most genome-wide association and fine-mapping studies to date have been conducted in individuals of European descent, and genetic studies of populations of Hispanic/Latino and African ancestry are limited. In addition, these populations have more complex linkage disequilibrium structure. In order to better define the genetic architecture of these understudied populations, we leveraged >100,000 phased sequences available from deep-coverage whole genome sequencing through the multi-ethnic NHLBI Trans-Omics for Precision Medicine (TOPMed) program to impute genotypes into admixed African and Hispanic/Latino samples with genome-wide genotyping array data. We demonstrated that using TOPMed sequencing data as the imputation reference panel improves genotype imputation quality in these populations, which subsequently enhanced gene-mapping power for complex traits. For rare variants with minor allele frequency (MAF) < 0.5%, we observed a 2.3- to 6.1-fold increase in the number of well-imputed variants, with 11-34% improvement in average imputation quality, compared to the state-of-the-art 1000 Genomes Project Phase 3 and Haplotype Reference Consortium reference panels. Impressively, even for extremely rare variants with minor allele count <10 (including singletons) in the imputation target samples, average information content rescued was >86%. Subsequent association analyses of TOPMed reference panel-imputed genotype data with hematological traits (hemoglobin (HGB), hematocrit (HCT), and white blood cell count (WBC)) in ~21,600 African-ancestry and ~21,700 Hispanic/Latino individuals identified associations with two rare variants in the HBB gene (rs33930165 with higher WBC [p = 8.8x10-15] in African populations, rs11549407 with lower HGB [p = 1.5x10-12] and HCT [p = 8.8x10-10] in Hispanics/Latinos). By comparison, neither variant would have been genome-wide significant if either 1000 Genomes Project Phase 3 or Haplotype Reference Consortium reference panels had been used for imputation. Our findings highlight the utility of the TOPMed imputation reference panel for identification of novel rare variant associations not previously detected in similarly sized genome-wide studies of under-represented African and Hispanic/Latino populations.
Conflict of interest statement
Edwin K Silverman and Michael H Cho have received grant support from GSK, MHC has received consulting fees from Genentech. Scott T. Weiss and Kathleen C. Barnes received royalties from UpToDate. Patrick T. Ellinor is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular diseases, and has also served on advisory boards or consulted for Bayer AG, Quest Diagnostics and Novartis. Steven A Lubitz receives sponsored research support from Bristol Myers Squibb / Pfizer, Bayer HealthCare, and Boehringer Ingelheim, and has consulted for Abbott, Quest Diagnostics, Bristol Myers Squibb / Pfizer. Other authors declared no conflicts of interest.
Figures
References
-
- Auer PL, Johnsen JM, Johnson AD, Logsdon BA, Lange LA, Nalls MA, et al. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am J Hum Genet. 2012;91(5):794–808. Epub 2012/10/30. 10.1016/j.ajhg.2012.08.031 . - DOI - PMC - PubMed
-
- Liu EY, Buyske S, Aragaki AK, Peters U, Boerwinkle E, Carlson C, et al. Genotype Imputation of MetabochipSNPs Using a Study-Specific Reference Panel of ~4,000 Haplotypes in African Americans From the Women’s Health Initiative. Genet Epidemiol. 2012;36(2):107–17. 10.1002/gepi.21603 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- N01 HC095160/HL/NHLBI NIH HHS/United States
- R01 HL118356/HL/NHLBI NIH HHS/United States
- UL1 RR033176/RR/NCRR NIH HHS/United States
- R01 HL112064/HL/NHLBI NIH HHS/United States
- HHSN268201100037C/HL/NHLBI NIH HHS/United States
- U01 HG007417/HG/NHGRI NIH HHS/United States
- HHSN268201100001I/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- R01 HL113323/HL/NHLBI NIH HHS/United States
- HHSN268201800012I/HB/NHLBI NIH HHS/United States
- R01 HL139731/HL/NHLBI NIH HHS/United States
- HHSN268201300026C/HL/NHLBI NIH HHS/United States
- RC2 AG036607/AG/NIA NIH HHS/United States
- HHSN268201800012C/HL/NHLBI NIH HHS/United States
- U01 HG007419/HG/NHGRI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- R01 HL104135/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL071251/HL/NHLBI NIH HHS/United States
- R01 HL123915/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01 HL087698/HL/NHLBI NIH HHS/United States
- K24 HL105780/HL/NHLBI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R01 HL121007/HL/NHLBI NIH HHS/United States
- R01 HL071259/HL/NHLBI NIH HHS/United States
- R01 HG006703/HG/NHGRI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- R01 HL141826/HL/NHLBI NIH HHS/United States
- HHSN268201800014I/HB/NHLBI NIH HHS/United States
- HHSN268201100004I/HL/NHLBI NIH HHS/United States
- N01 HC065233/HL/NHLBI NIH HHS/United States
- U01 HG007416/HG/NHGRI NIH HHS/United States
- T32 ES007018/ES/NIEHS NIH HHS/United States
- N01 HC065236/HL/NHLBI NIH HHS/United States
- R01 AR048797/AR/NIAMS NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- N01 HC065235/HL/NHLBI NIH HHS/United States
- R21 HL091397/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- HHSN268201100046C/HL/NHLBI NIH HHS/United States
- R01 HL129132/HL/NHLBI NIH HHS/United States
- R01 HL071250/HL/NHLBI NIH HHS/United States
- R01 HL142302/HL/NHLBI NIH HHS/United States
- R01 EY027004/EY/NEI NIH HHS/United States
- U01 HG007376/HG/NHGRI NIH HHS/United States
- 18SFRN34110082/AHA/American Heart Association-American Stroke Association/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- HHSN268201100003C/WH/WHI NIH HHS/United States
- R01 NS058700/NS/NINDS NIH HHS/United States
- HHSN268201300025C/HL/NHLBI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- F32 HL085989/HL/NHLBI NIH HHS/United States
- HHSN268201800014C/HL/NHLBI NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- R01 HL128914/HL/NHLBI NIH HHS/United States
- R01 HL119443/HL/NHLBI NIH HHS/United States
- HHSN268201200008C/HL/NHLBI NIH HHS/United States
- R37 HL066289/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- T32 HL129982/HL/NHLBI NIH HHS/United States
- R01 HL067348/HL/NHLBI NIH HHS/United States
- R01 HL113326/HL/NHLBI NIH HHS/United States
- HHSN268201300027C/HL/NHLBI NIH HHS/United States
- N01 HC065234/HL/NHLBI NIH HHS/United States
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 HL071051/HL/NHLBI NIH HHS/United States
- R01 DK101855/DK/NIDDK NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201800013I/MD/NIMHD NIH HHS/United States
- HHSN268201300028C/HL/NHLBI NIH HHS/United States
- HHSN271201100004C/AG/NIA NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- HHSN268200900041C/HL/NHLBI NIH HHS/United States
- HHSN268201500015C/HL/NHLBI NIH HHS/United States
- R56 HG010297/HG/NHGRI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- R01 DK116738/DK/NIDDK NIH HHS/United States
- M01 RR000052/RR/NCRR NIH HHS/United States
- HHSN268201100002C/WH/WHI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- R01 HL104608/HL/NHLBI NIH HHS/United States
- R01 AI132476/AI/NIAID NIH HHS/United States
- U01 HL072518/HL/NHLBI NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- HHSN268201800011C/HL/NHLBI NIH HHS/United States
- HHSN268201500014C/HL/NHLBI NIH HHS/United States
- M01 RR007122/RR/NCRR NIH HHS/United States
- R01 NS075107/NS/NINDS NIH HHS/United States
- R01 HL146500/HL/NHLBI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- R01 HL087263/HL/NHLBI NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- U01 HL072507/HL/NHLBI NIH HHS/United States
- R01 DK071891/DK/NIDDK NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- K01 HL130609/HL/NHLBI NIH HHS/United States
- R01 HL085571/HL/NHLBI NIH HHS/United States
- HHSN268201800015I/HB/NHLBI NIH HHS/United States
- R01 HL071205/HL/NHLBI NIH HHS/United States
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- T32 HL007284/HL/NHLBI NIH HHS/United States
- HHSN268201100003I/HL/NHLBI NIH HHS/United States
- HHSN268201100002I/HL/NHLBI NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201800010I/HB/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- HHSN268201300029C/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
- R01 HL090682/HL/NHLBI NIH HHS/United States
- HHSN268201800011I/HB/NHLBI NIH HHS/United States
- N01 HC065237/HL/NHLBI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- HHSN268201200008I/HL/NHLBI NIH HHS/United States
- R01 HL071258/HL/NHLBI NIH HHS/United States
- HHSN268201100001C/WH/WHI NIH HHS/United States
- R01 HL055673/HL/NHLBI NIH HHS/United States
- HHSN268201100004C/WH/WHI NIH HHS/United States
- R01 HL092301/HL/NHLBI NIH HHS/United States

