Investigation of somatic CNVs in brains of synucleinopathy cases using targeted SNCA analysis and single cell sequencing
- PMID: 31870437
- PMCID: PMC6929293
- DOI: 10.1186/s40478-019-0873-5
Investigation of somatic CNVs in brains of synucleinopathy cases using targeted SNCA analysis and single cell sequencing
Abstract
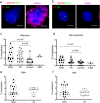
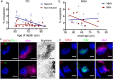
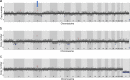
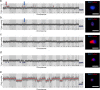
Synucleinopathies are mostly sporadic neurodegenerative disorders of partly unexplained aetiology, and include Parkinson's disease (PD) and multiple system atrophy (MSA). We have further investigated our recent finding of somatic SNCA (α-synuclein) copy number variants (CNVs, specifically gains) in synucleinopathies, using Fluorescent in-situ Hybridisation for SNCA, and single-cell whole genome sequencing for the first time in a synucleinopathy. In the cingulate cortex, mosaicism levels for SNCA gains were higher in MSA and PD than controls in neurons (> 2% in both diseases), and for MSA also in non-neurons. In MSA substantia nigra (SN), we noted SNCA gains in > 3% of dopaminergic (DA) neurons (identified by neuromelanin) and neuromelanin-negative cells, including olig2-positive oligodendroglia. Cells with CNVs were more likely to have α-synuclein inclusions, in a pattern corresponding to cell categories mostly relevant to the disease: DA neurons in Lewy-body cases, and other cells in the striatonigral degeneration-dominant MSA variant (MSA-SND). Higher mosaicism levels in SN neuromelanin-negative cells may correlate with younger onset in typical MSA-SND, and in cingulate neurons with younger death in PD. Larger sample sizes will, however, be required to confirm these putative findings. We obtained genome-wide somatic CNV profiles from 169 cells from the substantia nigra of two MSA cases, and pons and putamen of one. These showed somatic CNVs in ~ 30% of cells, with clonality and origins in segmental duplications for some. CNVs had distinct profiles based on cell type, with neurons having a mix of gains and losses, and other cells having almost exclusively gains, although control data sets will be required to determine possible disease relevance. We propose that somatic SNCA CNVs may contribute to the aetiology and pathogenesis of synucleinopathies, and that genome-wide somatic CNVs in MSA brain merit further study.
Keywords: Alpha-synuclein; Mosaicism; Multiple system atrophy; Parkinson’s disease; SNCA; Single cell sequencing; Somatic mutation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Somatic copy number gains of α-synuclein (SNCA) in Parkinson's disease and multiple system atrophy brains.Brain. 2018 Aug 1;141(8):2419-2431. doi: 10.1093/brain/awy157. Brain. 2018. PMID: 29917054
-
Somatic SNCA Copy Number Variants in Multiple System Atrophy are Related to Pathology and Inclusions.Mov Disord. 2023 Feb;38(2):338-342. doi: 10.1002/mds.29291. Epub 2022 Nov 30. Mov Disord. 2023. PMID: 36448620
-
Genetics of Synucleinopathies.Cold Spring Harb Perspect Med. 2018 Jun 1;8(6):a024109. doi: 10.1101/cshperspect.a024109. Cold Spring Harb Perspect Med. 2018. PMID: 28213435 Free PMC article. Review.
-
Somatic CNV Detection by Single-Cell Whole-Genome Sequencing in Postmortem Human Brain.Methods Mol Biol. 2023;2561:205-230. doi: 10.1007/978-1-0716-2655-9_11. Methods Mol Biol. 2023. PMID: 36399272
-
Somatic mutations in neurodegeneration: An update.Neurobiol Dis. 2020 Oct;144:105021. doi: 10.1016/j.nbd.2020.105021. Epub 2020 Jul 24. Neurobiol Dis. 2020. PMID: 32712267 Review.
Cited by
-
Multiple system atrophy.Nat Rev Dis Primers. 2022 Aug 25;8(1):56. doi: 10.1038/s41572-022-00382-6. Nat Rev Dis Primers. 2022. PMID: 36008429 Review.
-
Single-cell somatic copy number variants in brain using different amplification methods and reference genomes.Commun Biol. 2024 Oct 9;7(1):1288. doi: 10.1038/s42003-024-06940-w. Commun Biol. 2024. PMID: 39384904 Free PMC article.
-
A technology of a different sort: microraft arrays.Lab Chip. 2021 Sep 7;21(17):3204-3218. doi: 10.1039/d1lc00506e. Epub 2021 Aug 4. Lab Chip. 2021. PMID: 34346456 Free PMC article. Review.
-
Infectious origin of Alzheimer's disease: Amyloid beta as a component of brain antimicrobial immunity.PLoS Pathog. 2022 Nov 17;18(11):e1010929. doi: 10.1371/journal.ppat.1010929. eCollection 2022 Nov. PLoS Pathog. 2022. PMID: 36395147 Free PMC article. Review.
-
Tracing brain genotoxic stress in Parkinson's disease with a novel single-cell genetic sensor.Sci Adv. 2022 Apr 15;8(15):eabd1700. doi: 10.1126/sciadv.abd1700. Epub 2022 Apr 15. Sci Adv. 2022. PMID: 35427151 Free PMC article.
References
-
- Alegre-Abarrategui Javier, Brimblecombe Katherine R., Roberts Rosalind F., Velentza-Almpani Elisavet, Tilley Bension S., Bengoa-Vergniory Nora, Proukakis Christos. Selective vulnerability in α-synucleinopathies. Acta Neuropathologica. 2019;138(5):681–704. doi: 10.1007/s00401-019-02010-2. - DOI - PMC - PubMed
-
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, Warren WC, Magrini V, McGrath SD, Li YI, Wilson RK, Eichler EE. Characterizing the major structural variant alleles of the human genome. Cell. 2019;176:663–675.e19. doi: 10.1016/j.cell.2018.12.019. - DOI - PMC - PubMed
-
- Bae T, Tomasini L, Mariani J, Zhou B, Roychowdhury T, Franjic D, Pletikos M, Pattni R, Chen B-J, Venturini E, Riley-Gillis B, Sestan N, Urban AE, Abyzov A, Vaccarino FM. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2017;555(80):eaan8690. doi: 10.1126/science.aan8690. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT093205MA/WT_/Wellcome Trust/United Kingdom
- G0802760/MRC_/Medical Research Council/United Kingdom
- MR/J004758/1/MRC_/Medical Research Council/United Kingdom
- Department of Health [UK]/International
- FC001202/MRC_/Medical Research Council/United Kingdom
- G1001253/MRC_/Medical Research Council/United Kingdom
- 8811.2/Michael J. Fox Foundation for Parkinson's Research/International
- FC001202/WT_/Wellcome Trust/United Kingdom
- WT104033/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- FC001202/CRUK_/Cancer Research UK/United Kingdom
- MR/S01165X/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

