Clinical Mutations That Partially Activate the Stringent Response Confer Multidrug Tolerance in Staphylococcus aureus
- PMID: 31871080
- PMCID: PMC7038295
- DOI: 10.1128/AAC.02103-19
Clinical Mutations That Partially Activate the Stringent Response Confer Multidrug Tolerance in Staphylococcus aureus
Abstract
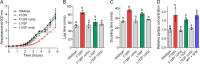
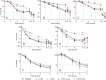
Antibiotic tolerance is an underappreciated antibiotic escape strategy that is associated with recurrent and relapsing infections, as well as acting as a precursor to resistance. Tolerance describes the ability of a bacterial population to survive transient exposure to an otherwise lethal concentration of antibiotic without exhibiting an elevated MIC. It is detected in time-kill assays as a lower rate of killing than a susceptible strain and can be quantified by the metric minimum duration for killing (MDK). The molecular mechanisms behind tolerance are varied, but activation of the stringent response (SR) via gene knockouts and/or chemical induction has long been associated with tolerance. More recently, two Gram-positive clinical isolates from persistent bacteremias were found to bear mutations in the SR controller, Rel, that caused elevated levels of the alarmone (p)ppGpp. Here, we show that introduction of either of these mutations into Staphylococcus aureus confers tolerance to five different classes of antibiotic as a result of (p)ppGpp-mediated growth defects (longer lag time and/or lower growth rate). The degree of tolerance is related to the severity of the growth defect and ranges from a 1.5- to 3.1-fold increase in MDK. Two classes of proposed SR inhibitor were unable to reverse or reduce this tolerance. Our findings reveal the significance of SR-activating mutations in terms of tolerance and clinical treatment failures. The panel of strains reported here provide a clinically relevant model of tolerance for further investigation of its link to resistance development, as well as potential validation of high-throughput tolerance screens.
Keywords: Staphylococcus aureus; antibiotic tolerance; minimum duration for killing; stringent response; time-kill curves.
Copyright © 2020 American Society for Microbiology.
Figures


References
-
- Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
-
- World Health Organization. 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland: https://www.who.int/antimicrobial-resistance/publications/global-action-....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

