Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself
- PMID: 31871199
- PMCID: PMC6955303
- DOI: 10.1073/pnas.1914072117
Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself
Abstract
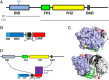
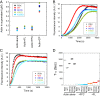
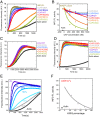
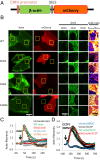
INF2 is a formin protein that accelerates actin polymerization. A common mechanism for formin regulation is autoinhibition, through interaction between the N-terminal diaphanous inhibitory domain (DID) and C-terminal diaphanous autoregulatory domain (DAD). We recently showed that INF2 uses a variant of this mechanism that we term "facilitated autoinhibition," whereby a complex consisting of cyclase-associated protein (CAP) bound to lysine-acetylated actin (KAc-actin) is required for INF2 inhibition, in a manner requiring INF2-DID. Deacetylation of actin in the CAP/KAc-actin complex activates INF2. Here we use lysine-to-glutamine mutations as acetylmimetics to map the relevant lysines on actin for INF2 regulation, focusing on K50, K61, and K328. Biochemically, K50Q- and K61Q-actin, when bound to CAP2, inhibit full-length INF2 but not INF2 lacking DID. When not bound to CAP, these mutant actins polymerize similarly to WT-actin in the presence or absence of INF2, suggesting that the effect of the mutation is directly on INF2 regulation. In U2OS cells, K50Q- and K61Q-actin inhibit INF2-mediated actin polymerization when expressed at low levels. Direct-binding studies show that the CAP WH2 domain binds INF2-DID with submicromolar affinity but has weak affinity for actin monomers, while INF2-DAD binds CAP/K50Q-actin 5-fold better than CAP/WT-actin. Actin in complex with full-length CAP2 is predominately ATP-bound. These interactions suggest an inhibition model whereby CAP/KAc-actin serves as a bridge between INF2 DID and DAD. In U2OS cells, INF2 is 90-fold and 5-fold less abundant than CAP1 and CAP2, respectively, suggesting that there is sufficient CAP for full INF2 inhibition.
Keywords: U2OS; WH2 motif; cyclase-associated protein; mitochondria; nucleation.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Higgs H. N., Formin proteins: A domain-based approach. Trends Biochem. Sci. 30, 342–353 (2005). - PubMed
-
- Goode B. L., Eck M. J., Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 (2007). - PubMed
-
- Li F., Higgs H. N., The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

