Bat-Borne Influenza A Viruses: An Awakening
- PMID: 31871229
- PMCID: PMC7849349
- DOI: 10.1101/cshperspect.a038612
Bat-Borne Influenza A Viruses: An Awakening
Abstract
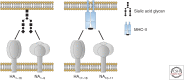
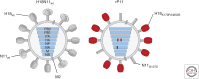
Influenza A viruses (IAVs) originating from aquatic waterfowl recurrently cross interspecies barriers, which is greatly facilitated by utilizing cell surface-exposed monosaccharide sialic acids located on vertebrate cells as a universal host cell receptor. These glycan structures are first bound by the viral hemagglutinin (HA) for cell entry and then cleaved by the viral neuraminidase (NA) for particle release. In contrast, viruses of the recently identified bat-borne IAV subtypes H17N10 and H18N11 encode HA and NA homologs unable to interact with sialic acid residues despite a high degree of structural homology with their conventional counterparts. However, the most recent findings show that bat IAV HAs make use of the major histocompatibility complex class II proteins of different vertebrate species to gain entry into host cells, potentially permitting a broader host tropism. This review recapitulates current progress in the field of bat IAV research including the first assessment of the spillover potential of these bat viruses into other mammals.
Copyright © 2021 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
-
- Ann J, Abed Y, Beaulieu E, Bouhy X, Joly MH, Dubé K, Carbonneau J, Hamelin ME, Mallett C, Boivin G. 2016. Impact of a large deletion in the neuraminidase protein identified in a laninamivir-selected influenza A/Brisbane/10/2007 (H3N2) variant on viral fitness in vitro and in ferrets. Influenza Other Respir Viruses 10: 122–126. 10.1111/irv.12356 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
