Structure and Function of the Influenza Virus Transcription and Replication Machinery
- PMID: 31871230
- PMCID: PMC7334866
- DOI: 10.1101/cshperspect.a038398
Structure and Function of the Influenza Virus Transcription and Replication Machinery
Abstract
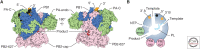
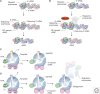
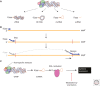
Transcription and replication of the influenza virus RNA genome is catalyzed by the viral heterotrimeric RNA-dependent RNA polymerase in the context of viral ribonucleoprotein (vRNP) complexes. Atomic resolution structures of the viral RNA synthesis machinery have offered insights into the initiation mechanisms of viral transcription and genome replication, and the interaction of the viral RNA polymerase with host RNA polymerase II, which is required for the initiation of viral transcription. Replication of the viral RNA genome by the viral RNA polymerase depends on host ANP32A, and host-specific sequence differences in ANP32A underlie the poor activity of avian influenza virus polymerases in mammalian cells. A failure to faithfully copy the viral genome segments can lead to the production of aberrant viral RNA products, such as defective interfering (DI) RNAs and mini viral RNAs (mvRNAs). Both aberrant RNA types have been implicated in innate immune responses against influenza virus infection. This review discusses recent insights into the structure-function relationship of the viral RNA polymerase and its role in determining host range and virulence.
Copyright © 2020 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
