The Ecology and Evolution of Influenza Viruses
- PMID: 31871237
- PMCID: PMC7328453
- DOI: 10.1101/cshperspect.a038489
The Ecology and Evolution of Influenza Viruses
Abstract
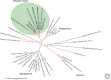
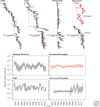
The patterns and processes of influenza virus evolution are of fundamental importance, underpinning such traits as the propensity to emerge in new host species and the ability to rapidly generate antigenic variation. Herein, we review key aspects of the ecology and evolution of influenza viruses. We begin with an exploration of the origins of influenza viruses within the orthomyxoviruses, showing how our perception of the evolutionary history of these viruses has been transformed with metagenomic sequencing. We then outline the diversity of virus subtypes in different species and the processes by which these viruses have emerged in new hosts, with a particular focus on the role played by segment reassortment. We then turn our attention to documenting the spread and phylodynamics of seasonal influenza A and B viruses in human populations, including the drivers of antigenic evolution, and finish with a discussion of virus diversity and evolution at the scale of individual hosts.
Copyright © 2020 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
