Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital PCR
- PMID: 31872202
- PMCID: PMC7250044
- DOI: 10.1039/c9lc01212e
Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital PCR
Abstract
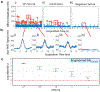
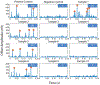
Sepsis due to antimicrobial resistant pathogens is a major health problem worldwide. The inability to rapidly detect and thus treat bacteria with appropriate agents in the early stages of infections leads to excess morbidity, mortality, and healthcare costs. Here we report a rapid diagnostic platform that integrates a novel one-step blood droplet digital PCR assay and a high throughput 3D particle counter system with potential to perform bacterial identification and antibiotic susceptibility profiling directly from whole blood specimens, without requiring culture and sample processing steps. Using CTX-M-9 family ESBLs as a model system, we demonstrated that our technology can simultaneously achieve unprecedented high sensitivity (10 CFU per ml) and rapid sample-to-answer assay time (one hour). In head-to-head studies, by contrast, real time PCR and BioRad ddPCR only exhibited a limit of detection of 1000 CFU per ml and 50-100 CFU per ml, respectively. In a blinded test inoculating clinical isolates into whole blood, we demonstrated 100% sensitivity and specificity in identifying pathogens carrying a particular resistance gene. We further demonstrated that our technology can be broadly applicable for targeted detection of a wide range of antibiotic resistant genes found in both Gram-positive (vanA, nuc, and mecA) and Gram-negative bacteria, including ESBLs (blaCTX-M-1 and blaCTX-M-2 families) and CREs (blaOXA-48 and blaKPC), as well as bacterial speciation (E. coli and Klebsiella spp.) and pan-bacterial detection, without requiring blood culture or sample processing. Our rapid diagnostic technology holds great potential in directing early, appropriate therapy and improved antibiotic stewardship in combating bloodstream infections and antibiotic resistance.
Conflict of interest statement
Figures





References
-
- World Health Organization (WHO). Fact sheet: Sepsis [Internet]. [cited 2019 Mar 29]. Available from: https://www.who.int/news-room/fact-sheets/detail/sepsis
-
- The Alliance for the, Prudent Use of Antibiotics. The cost of antibiotic resistance to U.S. families and the health care system. 2010.
-
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006. June;34(6):1589–96. - PubMed
-
- Kang C-I, Kim S-H, Park WB, Lee K-D, Kim H-B, Kim E-C, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005. February;49(2):760–6. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

