Subcellular drug targeting illuminates local kinase action
- PMID: 31872801
- PMCID: PMC6930117
- DOI: 10.7554/eLife.52220
Subcellular drug targeting illuminates local kinase action
Abstract
Deciphering how signaling enzymes operate within discrete microenvironments is fundamental to understanding biological processes. A-kinase anchoring proteins (AKAPs) restrict the range of action of protein kinases within intracellular compartments. We exploited the AKAP targeting concept to create genetically encoded platforms that restrain kinase inhibitor drugs at distinct subcellular locations.
Keywords: biochemistry; cell biology; chemical biology; kinases; local signaling; mitosis; zebrafish.
© 2019, Bucko et al.
Conflict of interest statement
PB, CL, LR, IG, AB, LW, FS, DM, HH, JS No competing interests declared
Figures


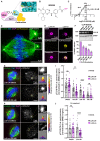


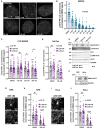

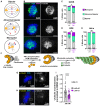


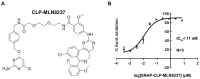









Comment in
-
Pharmacology on Target.Trends Pharmacol Sci. 2020 Apr;41(4):227-230. doi: 10.1016/j.tips.2020.02.002. Epub 2020 Feb 28. Trends Pharmacol Sci. 2020. PMID: 32122673 Free PMC article.
References
-
- Asteriti IA, Di Cesare E, De Mattia F, Hilsenstein V, Neumann B, Cundari E, Lavia P, Guarguaglini G. The Aurora-A inhibitor MLN8237 affects multiple mitotic processes and induces dose-dependent mitotic abnormalities and aneuploidy. Oncotarget. 2014;5:6229–6242. doi: 10.18632/oncotarget.2190. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32GM008268/NH/NIH HHS/United States
- R01 DK119192/DK/NIDDK NIH HHS/United States
- R01 GM086858/GM/NIGMS NIH HHS/United States
- R01 GM127621/GM/NIGMS NIH HHS/United States
- R01 GM069429/GM/NIGMS NIH HHS/United States
- R01GM127621/NH/NIH HHS/United States
- R01 DK105542/DK/NIDDK NIH HHS/United States
- R01 DK119186/DK/NIDDK NIH HHS/United States
- 1R01DK119192-01/NH/NIH HHS/United States
- R01GM086858/NH/NIH HHS/United States
- 5R01DK105542/NH/NIH HHS/United States
- DGE-1762114/National Science Foundation/International
- T32 GM008268/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

