Selecting Appropriate Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Studies in Isolated and Cultured Ocular Surface Epithelia
- PMID: 31873107
- PMCID: PMC6927975
- DOI: 10.1038/s41598-019-56054-1
Selecting Appropriate Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Studies in Isolated and Cultured Ocular Surface Epithelia
Abstract
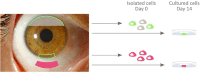
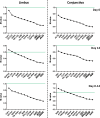
The introduction of tissue engineering has allowed scientists to push the boundaries and treat seriously damaged ocular surface epithelia. They have managed to do this through the development of biological substitutes that restore, maintain or improve tissue function. To ensure the generation of a therapeutically safe and effective graft, knowledge on the transcriptional profile of native and cultured ocular surface epithelia is of undeniable value. Gene expression studies are, however, only as reliable as their proper selection of internal reaction controls or reference genes. In this study, we determined the expression stability of a number of reference genes: 18s rRNA, ACTB, ATP5B, CyC1, EIF4A2, GAPDH, RPL13A, SDHA, TOP1, UBC, and YWHAZ in primary isolates as well as in ex vivo cultured ocular surface epithelia explants (day 0 and/or day 14). Expression stability of the reference genes was assessed with both the geNorm and NormFinder software that use a pairwise comparison and a model-based approach, respectively. Our results extend the general recommendation of using multiple reference genes for normalization purposes to our model systems and provide an overview of several references genes that are likely to be stable in similar culture protocols.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Pregnancy influences the selection of appropriate reference genes in mouse tissue: Determination of appropriate reference genes for quantitative reverse transcription PCR studies in tissues from the female mouse reproductive axis.Gene. 2021 Oct 30;801:145855. doi: 10.1016/j.gene.2021.145855. Epub 2021 Jul 20. Gene. 2021. PMID: 34293448
-
Variation in stability of housekeeping genes in healthy and adhesion-related mesothelium.Fertil Steril. 2012 Oct;98(4):1023-7. doi: 10.1016/j.fertnstert.2012.06.033. Epub 2012 Jul 13. Fertil Steril. 2012. PMID: 22795637
-
Identification of optimal reference genes for transcriptomic analyses in normal and diseased human heart.Cardiovasc Res. 2018 Feb 1;114(2):247-258. doi: 10.1093/cvr/cvx182. Cardiovasc Res. 2018. PMID: 29036603
-
Selection of suitable reference genes for normalization of quantitative real-time polymerase chain reaction in human cartilage endplate of the lumbar spine.PLoS One. 2014 Feb 18;9(2):e88892. doi: 10.1371/journal.pone.0088892. eCollection 2014. PLoS One. 2014. PMID: 24558443 Free PMC article.
-
Search for appropriate reference genes for quantitative reverse transcription PCR studies in somite, prosencephalon and heart of early mouse embryo.Gene. 2019 Aug 20;710:148-155. doi: 10.1016/j.gene.2019.05.042. Epub 2019 Jun 2. Gene. 2019. PMID: 31167115
Cited by
-
Immunocytochemical characterization of ex vivo cultured conjunctival explants; marker validation for the identification of squamous epithelial cells and goblet cells.Front Med (Lausanne). 2023 Feb 27;10:1024926. doi: 10.3389/fmed.2023.1024926. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36923014 Free PMC article.
-
The carbon source-dependent pattern of antimicrobial activity and gene expression in Pseudomonas donghuensis P482.Sci Rep. 2021 May 26;11(1):10994. doi: 10.1038/s41598-021-90488-w. Sci Rep. 2021. PMID: 34040089 Free PMC article.
-
Characterisation of Gel-Forming Mucins Produced In Vivo and In Ex Vivo Conjunctival Explant Cultures.Int J Mol Sci. 2021 Sep 29;22(19):10528. doi: 10.3390/ijms221910528. Int J Mol Sci. 2021. PMID: 34638869 Free PMC article.
-
Identification of an appropriate reference gene for normalization of qRT-PCR expression analyses in human breast cancer cell lines: application to L-arginine depletion studies.J Cancer Res Clin Oncol. 2025 Mar 26;151(3):122. doi: 10.1007/s00432-025-06165-2. J Cancer Res Clin Oncol. 2025. PMID: 40133575 Free PMC article.
-
Reference genes validation in tear fluid for RNA analysis in ocular surface disease.Sci Rep. 2025 Jul 9;15(1):24777. doi: 10.1038/s41598-025-05638-1. Sci Rep. 2025. PMID: 40634400 Free PMC article.
References
-
- Cieza, A., Keel, S., Kocur, I., Mccoy, M. & Mariotti, S. P. World report on vision. (World Health Organization, 2019).
-
- Thoft RA, Friend J. The X, Y, Z hypothesis of corneal epithelial maintenance. Invest Ophthalmol Vis Sci. 1983;24:1442–1443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

