A large self-transmissible resistance plasmid from Nigeria contains genes that ameliorate a carrying cost
- PMID: 31873110
- PMCID: PMC6927977
- DOI: 10.1038/s41598-019-56064-z
A large self-transmissible resistance plasmid from Nigeria contains genes that ameliorate a carrying cost
Abstract
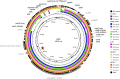
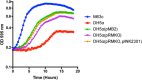
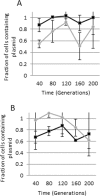
Antimicrobial resistance is rapidly expanding, in a large part due to mobile genetic elements. We screened 94 fecal fluoroquinolone-resistant Escherichia coli isolates from Nigeria for six plasmid-mediated quinolone resistance (PMQR) genes. Sixteen isolates harbored at least one of the PMQR genes and four were positive for aac-6-Ib-cr. In one strain, aac-6-Ib-cr was mapped to a 125 Kb self-transmissible IncFII plasmid, pMB2, which also bears blaCTX-M-15, seven other functional resistance genes and multiple resistance pseudogenes. Laboratory strains carrying pMB2 grew faster than isogenic strains lacking the plasmid in both rich and minimal media. We excised a 32 Kb fragment containing transporter genes and several open-reading frames of unknown function. The resulting 93 Kb mini-plasmid conferred slower growth rates and lower fitness than wildtype pMB2. Trans-complementing the deletion with the cloned sitABCD genes confirmed that they accounted for the growth advantage conferred by pMB2 in iron-depleted media. pMB2 is a large plasmid with a flexible resistance region that contains loci that can account for evolutionary success in the absence of antimicrobials. Ancillary functions conferred by resistance plasmids can mediate their retention and transmissibility, worsening the trajectory for antimicrobial resistance and potentially circumventing efforts to contain resistance through restricted use.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

