miRNA551b-3p Activates an Oncostatin Signaling Module for the Progression of Triple-Negative Breast Cancer
- PMID: 31875548
- PMCID: PMC7380555
- DOI: 10.1016/j.celrep.2019.11.085
miRNA551b-3p Activates an Oncostatin Signaling Module for the Progression of Triple-Negative Breast Cancer
Abstract
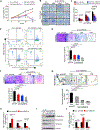
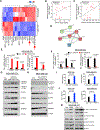
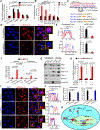
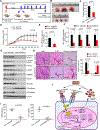
Genomic amplification of 3q26.2 locus leads to the increased expression of microRNA 551b-3p (miR551b-3p) in triple-negative breast cancer (TNBC). Our results demonstrate that miR551b-3p translocates to the nucleus with the aid of importin-8 (IPO8) and activates STAT3 transcription. As a consequence, miR551b upregulates the expression of oncostatin M receptor (OSMR) and interleukin-31 receptor-α (IL-31RA) as well as their ligands OSM and IL-31 through STAT3 transcription. We defined this set of genes induced by miR551b-3p as the "oncostatin signaling module," which provides oncogenic addictions in cancer cells. Notably, OSM is highly expressed in TNBC, and the elevated expression of OSM associates with poor outcome in estrogen-receptor-negative breast cancer patients. Conversely, targeting miR551b with anti-miR551b-3p reduced the expression of the OSM signaling module and reduced tumor growth, as well as migration and invasion of breast cancer cells.
Keywords: IPO8; OSMR; RNAi; STAT3; miR551b; miRNA-therapy; oncostatin.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures



References
-
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. (2007a). A module of negative feedback regulators defines growth factor signaling. Nat Genet 39, 503–512. - PubMed
-
- Anjali G, Kaur S, Lakra R, Taneja J, Kalsey GS, Nagendra A, Shrivastav TG, Devi MG, Malhotra N, Kriplani A, and Singh R (2015). FSH stimulates IRS-2 expression in human granulosa cells through cAMP/SP1, an inoperative FSH action in PCOS patients. Cell Signal 27, 2452–2466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA134682/CA/NCI NIH HHS/United States
- P50 CA217685/CA/NCI NIH HHS/United States
- R35 CA209904/CA/NCI NIH HHS/United States
- R01 CA123219/CA/NCI NIH HHS/United States
- UH3 TR000943/TR/NCATS NIH HHS/United States
- R01 HL112639/HL/NHLBI NIH HHS/United States
- P50 CA083639/CA/NCI NIH HHS/United States
- P50 CA058183/CA/NCI NIH HHS/United States
- R01 HL120585/HL/NHLBI NIH HHS/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- U01 CA217842/CA/NCI NIH HHS/United States
- R01 CA229907/CA/NCI NIH HHS/United States
- R01 HL123338/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

